
APA Style
Yi Wang, Xiushan Chen. (2025). Exploring Artesunate and Dihydroartemisinin for Mechanism on MMP-3 and MMP-13 via Molecular Docking and Molecular Dynamics. Molecular Modeling Connect, 2 (Article ID: 0006). https://doi.org/10.69709/MolModC.2025.142325MLA Style
Yi Wang, Xiushan Chen. "Exploring Artesunate and Dihydroartemisinin for Mechanism on MMP-3 and MMP-13 via Molecular Docking and Molecular Dynamics". Molecular Modeling Connect, vol. 2, 2025, Article ID: 0006, https://doi.org/10.69709/MolModC.2025.142325.Chicago Style
Yi Wang, Xiushan Chen. 2025. "Exploring Artesunate and Dihydroartemisinin for Mechanism on MMP-3 and MMP-13 via Molecular Docking and Molecular Dynamics." Molecular Modeling Connect 2 (2025): 0006. https://doi.org/10.69709/MolModC.2025.142325.
 ACCESS
Research Article
ACCESS
Research Article
Volume 2, Article ID: 2025.0006

Yi Wang
871916401@qq.com

Xiushan Chen
xiushanchen@126.com
1 Shandong Academy of Chinese Medicine, Jinan 250014, China
2 Haike Group Co., Ltd, Dongying 257000, China
* Author to whom correspondence should be addressed
Received: 16 Dec 2024 Accepted: 13 Apr 2025 Available Online: 22 Apr 2025 Published: 07 May 2025
The application of natural products has drawn increasing attention in recent times. Artesunate (ART) and Dihydroartemisinin (DHA), being natural products with anti-inflammatory and cartilage protective capabilities, have shown potential in osteoarthritis (OA) treatment. OA is a highly prevalent degenerative joint disease globally, leading to a diminished quality of life and is a major cause of disability. Matrix metalloproteinase 3 (MMP-3) and MMP-13 play crucial roles in OA progression. Although ART and DHA have been found to downregulate the protein levels of MMP-3 and MMP-13 and suppress the activity of inflammatory factors in chondrocytes, their interaction mechanisms with MMP-3 and MMP-13 remain unclear. This study aimed to conduct an in-silico exploration of the binding affinity, action mode, and stability of ART and DHA to the active sites of MMP-3 and MMP-13 using molecular docking and molecular dynamics simulation techniques. The results demonstrated that ART and DHA possess favorable binding affinities (≤−7.0 kcal/mol) for MMP-3 and MMP-13, establish hydrogen bonds and hydrophobic interactions with amino acid residues in the active sites, and form interactions with Zn2+. During the 50 ns simulation, the root mean square deviation (RMSD) of ART and DHA on MMP-3 was less than 2Å. The binding free energies were all negative, indicating a stable binding state. These findings disclose a novel function of ART and DHA as potential inhibitors of MMP-3 and MMP-13, presenting valuable insights and research directions for the development of novel OA therapeutics. However, further in vitro and in vivo investigations are required to validate the biological activities of these molecules.
Arthritis is a common clinical disease, with various subtypes, among which osteoarthritis (OA) is the most prevalent. OA is a chronic degenerative joint disease characterized by pain, stiffness, and limited mobility. Pathological features include degradation of articular cartilage extracellular matrix, synovial inflammation, osteophyte formation, joint space narrowing, ligament and meniscus damage, and subchondral bone remodelling, with irreversible degradation of the articular cartilage being the central pathological feature of OA [1,2,3]. OA is more common in middle-aged and older adults, and the exact cause is not fully understood [4]. In addition to ageing, the development of OA may be associated with multiple risk factors, including obesity, sex, injury, inflammation, and prolonged high-intensity exercise [5,6]. OA is estimated to affect hundreds of millions of people worldwide and has become a leading cause of disability [7], with a significant impact on the quality of daily life and a financial burden for patients [8]. In recent years, research on therapeutic agents for OA has made some progress. For instance, intra-articular injection of mesenchymal stem cells has significantly improved pain and function in patients with knee OA [9]. Additionally, a novel neurovascular inhibitory hydrogel has demonstrated potential in delaying the progression of OA by targeting inflammation and promoting cartilage repair [10]. However, the long-term effects of these treatments remain unclear, and there is heterogeneity among different research findings. As the world’s population ages and obesity rates rise, the prevalence of OA is expected to increase, so there is an urgent need for effective treatment strategies to relieve symptoms, slow disease progression, and improve patients’ quality of life [11,12]. Matrix Metalloproteinases (MMPs) are a class of Zn2+-dependent endopeptidases, and calcium, zinc, and other metal elements participate in the composition of these enzymes as cofactors. MMPs play a crucial role in the degradation and remodeling of the cartilage extracellular matrix, and they significantly impact the pathogenesis of OA. In particular, MMP-13 and MMP-3, both closely associated with the pathogenesis of osteoarthritis (OA), can degrade various matrix components, including proteoglycans, osteopontin, and type I, II, and III collagen, thereby playing key roles in the pathological progression of OA [13]. MMP-3, which is highly expressed in the early stages of OA, can degrade proteoglycans, thereby disrupting cartilage matrix integrity, and it can also activate other MMPs (such as MMP-1), further accelerating the degradation of the cartilage matrix [14,15]. MMP-13 is a key enzyme responsible for degrading type II collagen. It not only directly degrades collagen but also induces chondrocyte apoptosis, exacerbating cartilage damage [16]. Its expression persists throughout the entire progression of OA, and in the synovial tissues of OA patients, the high expression of MMP-13 is closely associated with inflammatory responses. Various inhibitors, such as AQU-019 [17] have been shown to effectively inhibit the activity of MMP-13 and demonstrate cartilage-protective effects in animal models. Therefore, MMP-3 and MMP-13 are potential targets for the development of OA therapeutic agents. Artesunate (ART) and Dihydroartemisinin (DHA) are natural compounds used to treat malaria and have various biological properties, including the regulation of inflammation and apoptosis in various cells. Several studies suggest that they play an important role in the treatment of OA. Yicheng Li et al. [18] found that ART delayed the progression of OA and significantly reduced the overexpression of MMP-3 and MMP-13 in chondrocyte-like ATDC 5 cells stimulated by IL-1β. Chengjin Zhao et al. [19] showed that ART ameliorated cartilage damage in OA, and that in in vitro cell experiments, ART reduced the expression of MMP-3 and MMP-13, increased type II collagen expression, and decreased the levels of IL-6 and TNF-α in cell supernatant. In addition, DHA has been shown to reduce OA by inhibiting aberrant bone remodeling and angiogenesis in subchondral bone, and decreasing MMP-13 expression in articular cartilage [20]. However, further clarification is still required regarding their interaction with MMP-3 and MMP-13. Computer simulation research is a new type of efficient, rapid, and economical means of drug discovery and mechanism of action research, which has been successfully applied to drug development [21,22,23,24]. Molecular docking, a computational technique for predicting protein-small molecule interactions, is pivotal in structure-based drug discovery. Recent advancements include EquiBind30 and TankBind29, leveraging deep learning for enhanced prediction accuracy of drug-protein binding structures [25,26]. DiffDock, employing a diffusion generative model, outperforms traditional docking algorithms [27]. FeatureDock, a transformer-based deep learning framework, utilizes protein local environment features to predict protein-ligand binding poses and facilitates virtual screening with robust scoring [28]. In OA-targeted drug research, molecular docking has elucidated binding mechanisms of OA-related drugs with their targets. For example, studies combining molecular docking and spectroscopy revealed the encapsulation and targeting mechanisms of camptothecin drugs with bovine serum albumin (BSA) [29]. Thus, docking simulations optimize drug binding to matrix metalloproteinases, facilitating the development of effective inhibitors. In this study, the interaction of MMP-3 and MMP-13 with ART and DHA was analyzed in detail by using molecular docking, and the molecular interaction pattern under physiological conditions was simulated at a dynamic level. The results showed that ART and DHA all exhibited significant binding affinity and stable binding to the protein, indicating their potential to inhibit MMP-3 and MMP-13. These results provide a scientific basis for understanding how ART and DHA work by reducing the protein expression of MMP-3 and MMP-13. Moreover, these findings provide an important reference point for the future discovery of more effective MMP-3 and MMP-13 inhibitors through structural modification, which has potentially important value in the prevention and treatment of arthritis. Nonetheless, further validation tests are required to confirm the biological activity and safety of these compounds.
2.1. Materials Molecular docking analysis was performed using Autodock Vina [30], PyMol [31], Discovery Studio 2024 client (DS), VMD and AutoDockTools-1.5.6 software [32]. Molecular dynamics (MD) simulation was carried out using Gromacs 5.1.4 software [33]. 2.2. Preparation of Ligand The 3D chemical structures of ART and DHA were originally obtained from the PubChem database (https://pubchem.ncbi.nlm.nih.gov (accessed on 6 July 2024)) and downloaded in SDF format. Subsequently, these compounds were processed using Pymol and saved in PDB format. To ensure that the ligands were properly prepared for subsequent molecular docking, they were optimized using AutoDockTools-1.5.6 before molecular docking. 2.3. Preparation of Protein The crystal structures of the MMPs are reported in the PDB (https://www.rcsb.org/), showing the protein conformation with co-crystallized with various inhibitors. In this study, crystal structures of MMP-3, MMP-13, along with their corresponding inhibitors, were used for analysis. The PDB IDs for MMP-3 and MMP-13 are 2D1O(A chain) [34] and 3KRY (A chain) [35], with resolution of 2.02 Å and 1.90Å, respectively. 2D1O file included one polypeptide chain (named chain A) with 171 residues, an inhibitor (PDB ID: FA4), three calcium ions, and two zinc ions. 3KRY file included one polypeptide chain (named chain A) with 164 residues, an inhibitor (PDB ID: 3KR), two calcium ions, and two zinc ions. To perform molecular docking studies on the selected crystal structures, water molecules and eutectic ligands were first removed, and hydrogen atoms were added to the protein structures. Both Zn2+ and Ca2+ are retained in the protein complex and are included in the analysis. 2.4. Molecular Docking After the preparations were completed, molecular docking was carried out using AutoDock Vina software to predict the binding pattern of the protein-ligand complex. PD-166793 is a potent, selective, orally active, and broad-spectrum inhibitor of MMP, exhibiting nanomolar potency against MMP-3 and MMP-13 (IC50 = 7 and 8 nM, respectively) [36]. Therefore, PD-166793, along with co-crystal ligands FA4 and 3KR, were adopted as positive controls. The active site of the protein structure was set as specific amino acid residues interacting with the cocrystallized ligand in X-ray crystallography. The grid parameters were as follows: the grid center coordinates of MMP-3 and MMP-13 are (29.194, 7.769, 14.716), (−11.439, −1.082, 0.865), respectively, and the grid sizes are both 23 Å. Protein-ligand interaction complexes were visualized using Pymol and DS to identify strong binding positions of active site residues and their binding distances. The conformation with the lowest binding energy and interaction with Zn2+ was selected for the initial structure of the subsequent MD simulations. 2.5. Molecular Dynamics To study their stability with MMP-3 and MMP-13, MD simulations of the optimal complex conformation were carried out for 50 ns using Gromacs 5.1.4 software. The simulation process included three stages: energy minimization, system balance, and free simulation. In the simulation, root mean square deviation (RMSD), root mean square fluctuation (RMSF), and binding free energy were used to evaluate the stability of the simulation and the binding strength of the protein-ligand complex. The RMSD is a measure of how much the protein conformation deviates from the initial structure in the simulation. A smaller value indicates that the simulated protein conformation is closer to the initial structure, allowing for an assessment of protein stability. When the RMSD value tends to stabilize, it indicates that the protein structure has reached a steady state, and the simulation can be considered to have stopped. However, RMSF analysis deeply reveals the internal dynamics of proteins, in particular the specific effects of ligand binding on the dynamics of different atoms or residues in MMP proteins. This analysis helps to identify protein regions that become more flexible or stable with ligands.
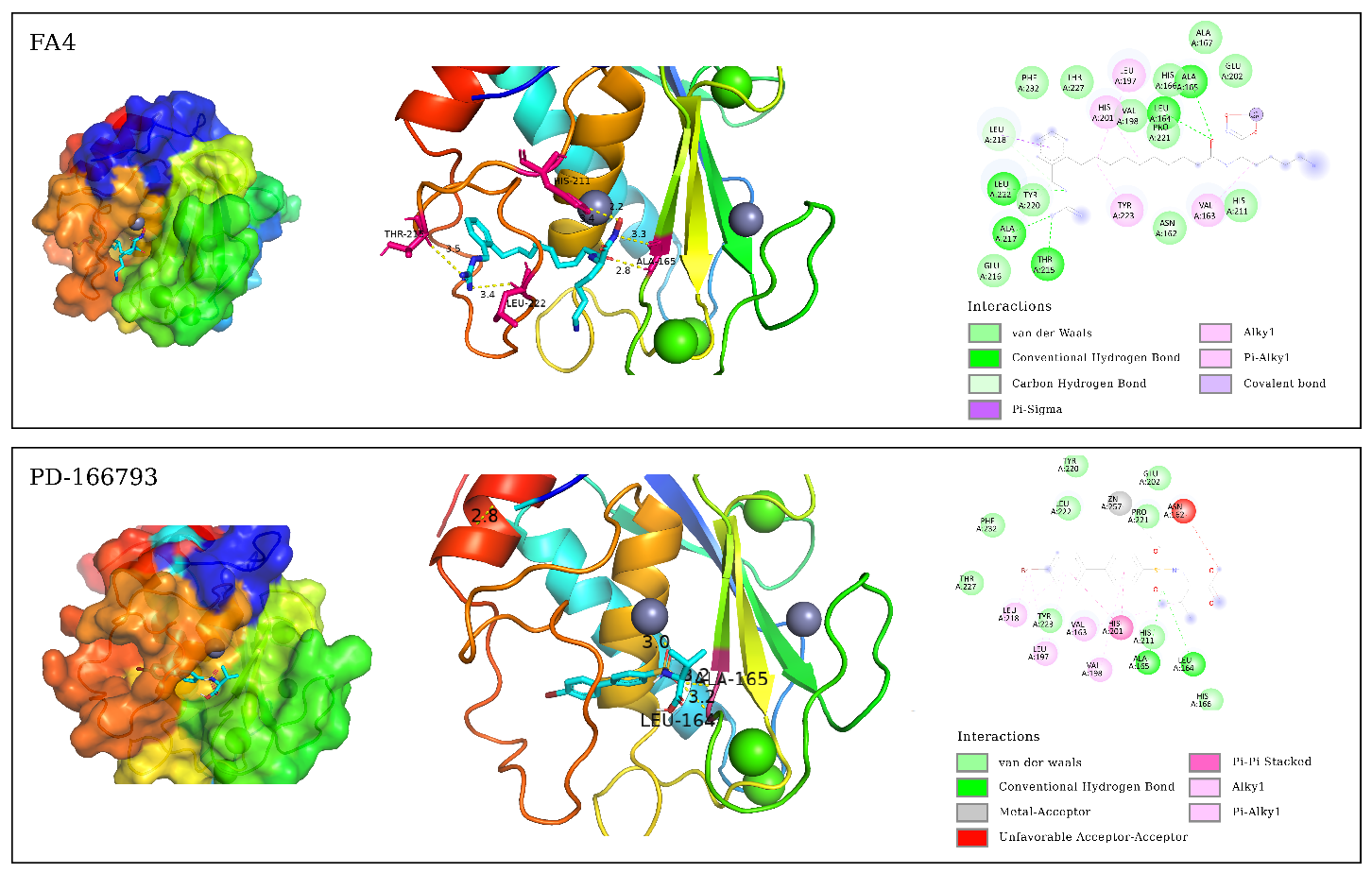
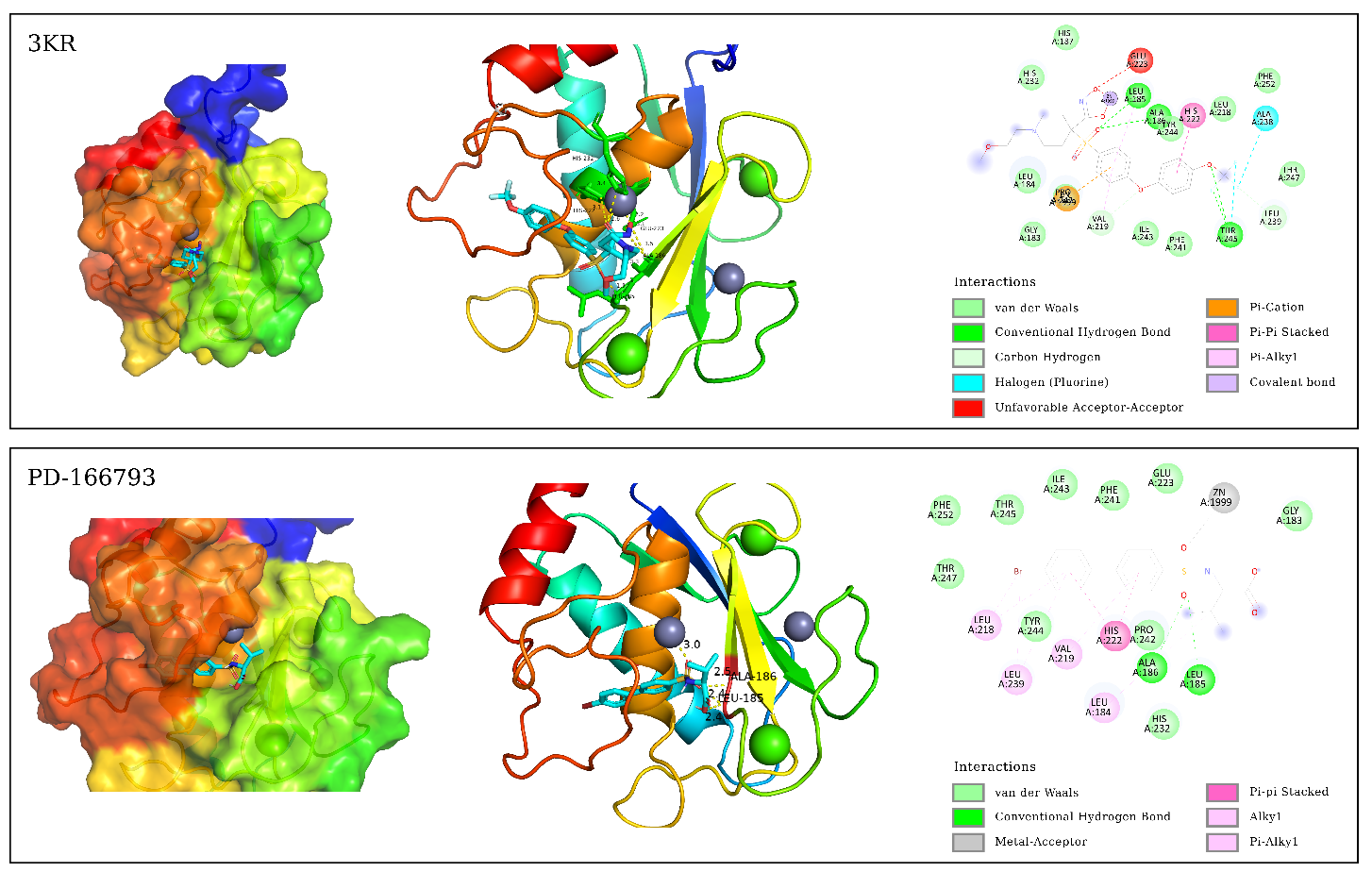
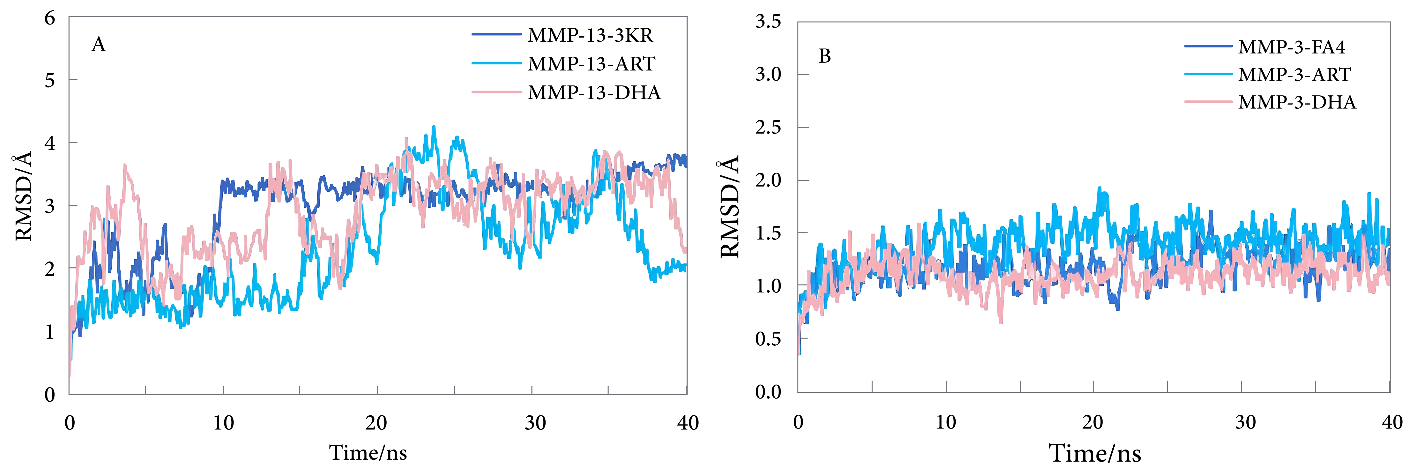
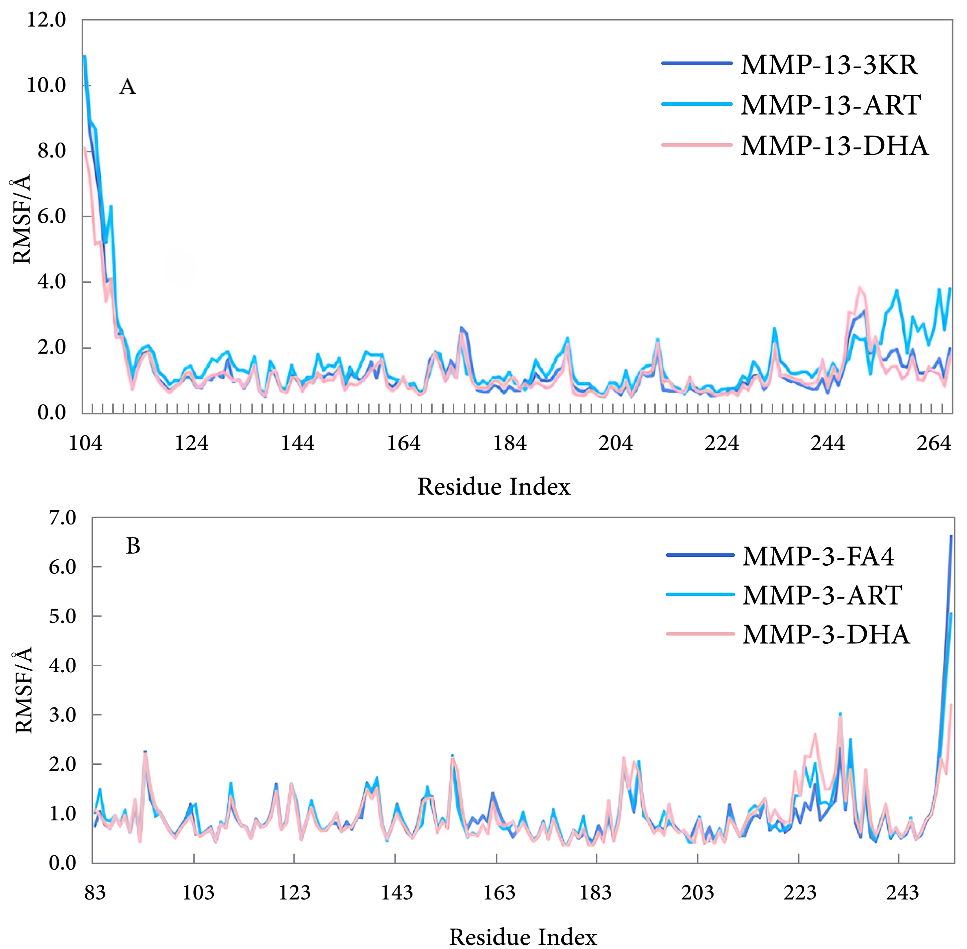
3.1. Binding Affinity Analysis Molecular docking technology evaluated the binding affinity of ART and DHA to the active site of MMP-3 and MMP-13. The docking results are shown in Table 1. The lower binding affinity value usually indicates a higher binding affinity, which means stronger interactions between the molecules. Binding affinity for docking with MMP-3, MMP-13. In this study, we evaluated the binding affinities of ART and DHA to MMP-3 and MMP-13 using molecular docking, and compared them with known inhibitors PD-166793, FA4, and 3KR. PD-166793 exhibited strong binding affinities of −9.2 kcal/mol for MMP-3 and −9.6 kcal/mol for MMP-13, which are consistent with its nanomolar potency (IC50 = 7 and 8 nM, respectively). These results validated the reliability of our docking method. FA4 and 3KR, which are co-crystallized ligands of MMP-3 and MMP-13, respectively, exhibited binding affinities of −7.8 kcal/mol and −9.5 kcal/mol to MMP-3 and MMP-13. ART demonstrated binding affinities of −8.1 kcal/mol for MMP-3 and −8.0 kcal/mol for MMP-13. which were comparable to those of FA4 for MMP-3 but slightly lower than those of 3KR for MMP-13. DHA showed a binding affinity of −7.6 kcal/mol for MMP-3, similar to FA4, but only −7.0 kcal/mol for MMP-13, indicating a significant difference from 3KR. Despite the lower binding affinities of ART and DHA compared to PD-166793, these compounds still exhibit substantial potential as inhibitors of MMP-3 and MMP-13. These findings suggested that ART may have dual inhibitory activity against both MMP-3 and MMP-13, while DHA appeared to be more selective for MMP-3. TIMP-3 [37], an endogenous inhibitor, had a binding affinity of approximately −7.8 kcal/mol for MMP-13. Additionally, the synthetic inhibitor Batimastat (BB-94), which had been extensively studied for its inhibitory effects on MMP-13, exhibited a binding affinity of around −9.0 kcal/mol [37]. In comparison, the binding affinities of ART and DHA were comparable to TIMP-3 but slightly lower than those of Batimastat. The recent studies demonstrated that ART and DHA can form covalent bonds with the zinc ions in the catalytic domain of MMPs, thereby showing strong inhibitory potential [38]. These results highlight the potential of ART and DHA as novel MMP inhibitors for the treatment of OA. 3.2. Binding Modes Analysis To further explore the binding modes of ART and DHA on MMP-3 and MMP-13, the interaction of the complex was visualized by Pymol and DS (Figure 1 and Figure 2). ART and DHA are located in the same active pocket as the co-crystal ligands (FA4 and 3KR), form interactions with the Zn2+ in MMP-3 and MMP-13, and have hydrogen bonds and hydrophobic interactions between the proteins. Figure 1 demonstrates the binding details of all compounds with MMP-3. FA4 formed a series of key non-covalent interaction networks with the active site residues of MMP-3, including the formation of six hydrogen bonds and five hydrophobic interactions with His211, Thr215, Leu222, Ala165, Leu164, Leu218, Ala217, Leu197, His201, Tyr223, and Val163. Furthermore, FA4 formed stable chelating interactions with Zn2+ within the active site through potential coordination atoms in its molecular structure. The PD-166793 formed two conventional hydrogen bonds with Leu164 and Ala165, and multiple hydrophobic interactions with Leu218, Leu197, Val163, Val198, and His201. Additionally, it coordinated with Zn2+, which is crucial for the activity of the enzyme. However, there was also an unfavorable factor present. The binding of ART on MMP-3 exhibited three hydrogen bonds and eight hydrophobic interactions with residues such as Ala167, His166, Ala165, Leu164, Val198, Leu222, His211, and Pro221. In particular, specific functional groups of ART interacted with Zn2+, further enhancing its binding affinity to MMP-3. Although DHA formed only one hydrogen bond interaction in binding to MMP-3, its five hydrophobic interactions with residues Leu164, Val198, Val163, and Pro221 also contributed significantly to the binding affinity. Figure 2 demonstrates the interaction of all compounds with MMP-13. There were six hydrogen bonds and five hydrophobic interactions between 3KR and residues His222, Leu185, Ala186, Thr245, Leu239, Val219, Ala238, Pro242, and Zn2+, but one interaction that may adversely affect binding was also observed. PD-166793 formed two conventional hydrogen bonds with LEU185 and ALA186, and established eight hydrophobic interactions with Leu218, Leu239, ValL219, Leu184, and His222. Additionally, it also coordinated with Zn2+. ART formed three hydrogen bonds and five hydrophobic interactions with Leu185, Leu239, Pro242, Ala186, His222, His226, and His232, while DHA formed two hydrogen bonds and five hydrophobic interactions with Leu185, Pro242, His232, Leu184, and Val219. In addition to the hydrogen bonds and hydrophobic interactions, the ligand coordinates with Zn2+, which is crucial for the enzyme’s activity. This coordination with Zn2+ is a key feature of effective MMP inhibitors, as Zn2+ plays a central role in the catalytic mechanism of these enzymes. Compared with known inhibitors such as PD-166793, which also coordinates with Zn2+, ART and DHA demonstrated a similar mode of interaction but with distinct binding affinities. All of them form interactions with Zn2+ through oxygen atoms. The bond lengths of ART and Zn2+ were 2.6 Å (MMP-3) and 3.1 Å (MMP-13), respectively, while the corresponding bond lengths of DHA were 3.4 Å (MMP-3) and 3.1 Å (MMP-13). These distance data provided important information for understanding their binding mode and possible inhibition mechanisms. 3.3. MD Simulation Analysis To study the dynamic behaviour and stability of bioactive compounds at the protein binding sites of MMP-3 and MMP-13, MD simulations were performed. Due to the limitations of our current computational resources, we extended the MD simulation time to 50 ns. During the entire simulation, visualization with VMD revealed that the ligand-maintained interaction with Zn²⁺. Although this duration was relatively short, we conducted a detailed analysis of the stability of our system within this timeframe. 3.3.1. Root Mean Square Deviation Figure 3 shows the changes in RMSD of the simulation system with simulation time. The figure indicated that the RMSD change trend of FA4, ART, and DHA in the MMP-3 system was largely consistent, with the value fluctuating within a narrow range after 10 ns and approaching equilibrium. The fluctuation range is less than 2.0 Å. MMP-13-3KR entered a stationary state after 10 ns, with RMSD fluctuating less. In contrast, MMP-13-ART and MMP-13-DHA exhibited significant fluctuations in the 50 ns simulation, with an RMSD average value of 2.286 ± 0.760 Å and 2.876 ± 0.582 Å, respectively. These fluctuations were evident from the beginning of the simulation, indicating that these two complexes failed to reach a stable state in the solvent and did not achieve convergence. The instability could be due to interactions between ligands and MMP-13’s active site. During simulations, ART and DHA might not have formed stable bindings with MMP-13, causing significant conformational changes in the solvent. This is consistent with previous findings that the binding of certain ligands to MMP-13 can increase the conformational flexibility of its active site, thereby affecting overall stability [39]. Additionally, simulation time may also be a crucial factor affecting stability. While a 50 ns simulation partially captures the system’s dynamic behavior, longer simulation timescales could potentially reveal more stable conformations. Hence, future investigations will contemplate extending the simulation duration to more comprehensively evaluate the stability and convergence of these complexes. 3.3.2. Root Mean Square Fluctuation As illustrated in Figure 4, the RMSF of the amino acids was measured in the MD to gain further insight into the fluctuations of amino acids in each system. The RMSF curves of ART, DHA-conjugated MMP-3, and MMP-13 complex systems were similar to those of the protoprotein complex, indicating that the two small molecules have analogous binding modes with FA4 and 3KR. In the MMP-3 system, most amino acid residues exhibited fluctuations within a range of less than 2.0 Å, except for those in the terminal regions. In contrast, in the MMP-13 system, residues in the central region displayed minimal fluctuations. 3.4. Binding Free Energy For a more detailed analysis of the binding affinities, we selected the structures from the last 10 ns of the MD trajectory and performed MM/PBSA binding free energy calculations. The calculated binding free energy values were presented in Table 2. Binding free energy for molecules with MMP-3, MMP-13 (kcal/mol). ΔEvdw, Van der Waals Energy; ΔEele, Electrostatic Energy; ΔEGB, Polar Solvation Energy; ΔESA, Non-Polar Solvation Energy; ΔEbind, Binding Free Energy. Among the ligands, ART exhibited the strongest binding free energy to MMP-3, second only to DHA. DHA had the lowest binding free energy with MMP-13, which is consistent with the binding affinities observed in the molecular docking studies. Specifically, the negative electrostatic energy reflected favorable electrostatic interactions between the ligand and the protein, while the negative van der Waals energy highlighted the presence of attractive forces at the binding interface. Additionally, the negative non-polar solvation energy suggested that the solvation process enhances the stability of the ligand-protein complex. Among them, the contribution of electrostatic interactions was the largest, followed by van der Waals energy. Collectively, the electrostatic, van der Waals, and non-polar solvation energies were all negative, indicating that these interactions significantly contribute to the binding of the small molecule to the protein.
PubChem CID
Ligand Name
Binding Affinity (kcal/mol)
MMP-3
MMP-13
6914601
FA4
−7.8
/
49866490
3KR
/
−9.5
9887870
PD-166793
−9.2
−9.6
6917864
ART
−8.1
−8.0
3000518
DHA
−7.6
−7.0
Complex
ΔEvdw
ΔEele
ΔEGB
ΔESA
ΔEbind
ART-MMP-3
−35.47
−90.67
24.36
−4.98
−106.76
DHA-MMP-3
−31.08
−110.51
44.42
−5.32
−102.49
ART-MMP-13
−34.33
−113.47
50.75
−5.02
−102.07
DHA-MMP-13
−30.21
−101.87
46.69
−6.85
−92.24
OA is a common joint disease that mainly affects the articular cartilage, resulting in joint pain, swelling, and reduced mobility. Traditional treatment of OA is to relieve symptoms through the use of analgesics, NSAIDs, hyaluronic acid conjugates, or surgical procedures [40]. However, they can only control the symptoms of OA, do not reverse the damage to the joints, and are likely to cause some unexpected side effects [41]. Natural products as a source of drug discovery offer an alternative to avoiding the side effects of traditional therapeutic agents [42,43]. Exploring natural products for potential OA treatments is a promising direction. The pathogenesis of OA involves three key factors: cartilage degradation, synovial immune response, and pathological changes in subchondral bone [44]. The progressive loss of articular cartilage is a significant sign of OA. In OA tissue, the accumulation of glycans in cartilage and chondro-specific type II collagen, two types of ECM, can rupture significantly, leading to chronic pain and disability in patients. MMPs contain many subtypes, including MMP-1, MMP-2, MMP-3, MMP-8, MMP-9, MMP-10, MMP-13, and MMP-14, which are capable of degrading different collagen proteins and are closely associated with the development of OA, most of which are involved in extracellular matrix remodelling and articular cartilage destruction in OA [45]. Meanwhile, several studies have shown that ART and DHA can improve OA, reduce the expression of MMP-3 and MMP-13, and improve type II collagen expression. However, their mechanism of action for improving OA, especially with MMP-3 and MMP-13, is unclear. Therefore, MMP-3 and MMP-13 were selected as target proteins in this study, and the interaction between ART and the DHA and both targets was investigated through molecular docking and MD. In this study, the potential binding capacity of ART and DHA on both MMP-3 and MMP-13 was evaluated using molecular docking methods. The results showed that ART has a significant binding affinity for these two key MMPs, which is consistent with its potential role in cartilage protection and inflammation regulation. In particular, the binding energy of ART on MMP-3 was the same as that of MMP-13, which may hint at its specific mode of action on different MMPs. Although DHA showed similar binding ability to MMP-3 as a positive control, it was slightly weaker to MMP-13, suggesting that it may have selective inhibitory properties of MMP-3. Hydrogen bonding, hydrophobic interactions, and Zn2+ interactions form the basis of the interaction between the ligand and the active site of the enzyme. In this study, it was found that ART and DHA formed different numbers of hydrogen bonds and hydrophobic interactions with MMP-3 and MMP-13. However, only ART was linked to the catalytic zinc of MMP-3 via an oxygen atom in its side chain, while the others interacted with the catalytic zinc through the oxygen atom in the ring.. These interactions jointly determined the binding strength and specificity of ligands and enzymes. ART interacted slightly more with the two enzymes than DHA, and the mode of action was slightly different, which is consistent with the difference in binding energy. Moreover, the length of the Zn2+ coordination bond between ART and MMP-3 was longer than that of Zn2+ of MMP-13, while DHA formed longer coordination bonds with both MMP-3 and MMP-13, and this change may reflect the effect of micro-environment differences in the active sites of different MMP isoenzymes on coordination geometry. Considering the above results, it was speculated that ART and DHA may decrease MMP-3 and MMP-13 expression by inhibiting MMP-3 and MMP-13. To verify the kinetic stability of the complex system, MD simulations of 50 ns were performed. The RMSD fluctuation with simulation time indicates that the MMP-3 complex system can reach equilibrium after simulation, exhibiting high kinetic stability. In contrast, the MMP-13 complex system exhibited significant fluctuations during the simulation and is not stable in the solvent. In MMP-3 and MMP-13, the amino acid residues that interacted with the ligand and bind tightly had lower RMSF values with less fluctuation and the weakest thermal movement of the atoms, which enhanced the stability of the amino acid residues in this region. Conversely, ART and DHA exhibited greater stability in their binding to MMP-3. The calculation of binding free energy indicated that ART had a strong binding ability with MMP-3, which was consistent with the docking results. Among these interactions, electrostatic interactions were the major stabilizing factors. This study deepens the understanding of the potential applications of ART and DHA in the treatment of OA, especially in the reduction of MMP-3 and MMP-13 expression. These findings not only provide a molecular basis for the structural optimization of ART and DHA as inhibitors of MMP-3 and MMP-13, but also provide valuable information for the design of novel MMP-targeting drugs. Although these simulation results provide valuable preliminary insights, we currently lack experimental validation to support these findings, relying primarily on computer simulations. To enhance the credibility and scientific nature of our conclusions, future research should include experimental validation. For example, cell experiments or animal models could be used to verify the inhibitory effects of ART and DHA on MMP-3 and MMP-13.
Molecular docking results show that ART and DHA exhibit significant affinity for MMP-3 and MMP-13 through hydrogen bonding, hydrophobic, and Zn2+ coordination, and that ART has better binding affinity in MMP-3 than the cocrystallized ligand FA4. MD simulation results indicate that ART and DHA bind to the two proteins like the native protein complex, and ART and DHA can stably bind to MMP-3. However, it is important to note that these in silico findings are subject to the limitations inherent in computational models, which may not fully account for the dynamic nature of protein-ligand interactions. Furthermore, the generalizability of these results may be limited by the specific conditions of the simulations and the need for experimental validation to confirm the biological relevance. In summary, ART and DHA may reduce the expression of MMP-3 and MMP-13 by inhibiting their activity, but further in vitro and in vivo studies are required to validate these preliminary computational insights.
| ART | Artesunate |
| DHA | Dihydroartemisinin |
| DS | Discovery Studio 2024 Client |
| MD | Molecular Dynamics |
| MMPs | Matrix Metalloproteinases |
| OA | Osteoarthritis |
| RMSD | Root Mean Square Deviation |
| RMSF | Root Mean Square Fluctuation |
Y.W.: Conceptualization, Methodology, Formal analysis, Investigation, Resources, Data curation, Writing—original draft. X.C.: Writing—review & editing, Visualization, Supervision, Funding acquisition.
Data supporting the results of this study are available upon request from the corresponding author.
No consent for publication is required, as the manuscript does not involve any individual personal data, images, videos, or other materials that would necessitate consent.
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
This work was supported by the Shandong Province Medical and Health Science and Technology Project (Grant No.: (202413050572)). The funding agencies had no role in the study design, data collection and analysis, decision to publish, or preparation of the article.
The authors would like to thank the Shandong Province Medical and Health Science and Technology Project (202413050572) for its support.
[1] Peng, Z.; Sun, H.; Bunpetch, V.; Koh, Y.; Wen, Y.; Wu, D.; Ouyang, H. The Regulation of Cartilage Extracellular Matrix Homeostasis in Joint Cartilage Degeneration and Regeneration. Biomaterials 2021, 268. [PubMed] [CrossRef]
[2] Malemud, C.J. Biologic Basis of Osteoarthritis: State of the Evidence. Curr. Opin. Rheumatol. 2015, 27, 289–294. [CrossRef] [PubMed]
[3] Yuan, C.; Pan, Z.; Zhao, K.; Li, J.; Sheng, Z.; Yao, X.; Liu, H.; Zhang, X.; Yang, Y.; Yu, D.; et al. Classification of Four Distinct Osteoarthritis Subtypes with a Knee Joint Tissue Transcriptome Atlas. Bone Res. 2020, 8, 38. [CrossRef] [PubMed]
[4] Hall, M.; van der Esch, M.; Hinman, R.S.; Peat, G.; de Zwart, A.; Quicke, J.G.; Runhaar, J.; Knoop, J.; van der Leeden, M.; de Rooij, M.; et al. How Does Hip Osteoarthritis Differ from Knee Osteoarthritis?. Osteoarthritis Cartilage 2022, 30, 32–41. [CrossRef] [PubMed]
[5] Yao, Q.; Wu, X.; Tao, C.; Gong, W.; Chen, M.; Qu, M.; Zhong, Y.; He, T.; Chen, S.; Xiao, G. Osteoarthritis: Pathogenic Signaling Pathways and Therapeutic Targets. Signal Transduct. Target. Ther. 2023, 8, 56. [CrossRef] [PubMed]
[6] Nedunchezhiyan, U.; Varughese, I.; Sun, A.R.; Wu, X.; Crawford, R.; Prasadam, I. Obesity, Inflammation, and Immune System in Osteoarthritis. Front. Immunol. 2022, 13. [CrossRef] [PubMed]
[7] Shane Anderson, A.; Loeser, R.F. Why Is Osteoarthritis an Age-Related Disease?. Best Pract. Res. Clin. Rheumatol. 2010, 24, 15–26. [CrossRef]
[8] Disease, G.B.D.; Injury, I.; Prevalence, C. Global, Regional, and National Incidence, Prevalence, and Years Lived with Disability for 354 Diseases and Injuries for 195 Countries and Territories, 1990-2017: A Systematic Analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1789–1858. [CrossRef]
[9] Cao, M.; Ou, Z.; Sheng, R.; Wang, Q.; Chen, X.; Zhang, C.; Dai, G.; Wang, H.; Li, J.; Zhang, X.; et al. Efficacy and Safety of Mesenchymal Stem Cells in Knee Osteoarthritis: A Systematic Review and Meta-Analysis of Randomized Controlled Trials. Stem Cell Res. Ther. 2025, 16, 122. [CrossRef]
[10] Qin, W.; Ma, Z.; Bai, G.; Qin, W.; Li, L.; Hao, D.; Wang, Y.; Yan, J.; Han, X.; Niu, W.; et al. Neurovascularization Inhibiting Dual Responsive Hydrogel for Alleviating the Progression of Osteoarthritis. Nat. Commun. 2025, 16, 1390. [CrossRef] [PubMed]
[11] Wan, Y.; Li, W.; Liao, Z.; Yan, M.; Chen, X.; Tang, Z. Selective MMP-13 Inhibitors: Promising Agents for the Therapy of Osteoarthritis. Curr. Med. Chem. 2020, 27, 3753–3769. [CrossRef] [PubMed]
[12] Hunter, D.J. Osteoarthritis. Best Pract. Res. Clin. Rheumatol. 2011, 25, 801–814. [PubMed] [CrossRef]
[13] Shiomi, T.; Lemaitre, V.; D’Armiento, J.; Okada, Y. Matrix Metalloproteinases, a Disintegrin and Metalloproteinases, and a Disintegrin and Metalloproteinases with Thrombospondin Motifs in Non-Neoplastic Diseases. Pathol. Int. 2010, 60, 477–496. [CrossRef]
[14] Wan, J.; Zhang, G.; Li, X.; Qiu, X.; Ouyang, J.; Dai, J.; Min, S. Matrix Metalloproteinase 3: A Promoting and Destabilizing Factor in the Pathogenesis of Disease and Cell Differentiation. Front. Physiol. 2021, 12. [CrossRef] [PubMed]
[15] Zhu, L.; Ma, M.; Xu, W.; Xiong, H. Exploring the Structural Properties, Osteoarthritis-Relieving Effects and Potential Mechanisms of Peptides from Eggshell Membrane Byproduct. Food Biosci. 2025, 66. [CrossRef]
[16] Ashruf, O.S.; Ansari, M.Y. Natural Compounds: Potential Therapeutics for the Inhibition of Cartilage Matrix Degradation in Osteoarthritis. Life 2022, 13. [CrossRef]
[17] Bendele, A.M.; Neelagiri, M.; Neelagiri, V.; Sucholeiki, I. Development of a Selective Matrix Metalloproteinase 13 (MMP-13) Inhibitor for the Treatment of Osteoarthritis. Eur. J. Med. Chem. 2021, 224, 113666. [PubMed] [CrossRef]
[18] Li, Y.; Mu, W.; Ren, J.; Wuermanbieke, S.; Wahafu, T.; Ji, B.; Ma, H.; Amat, A.; Zhang, K.; Cao, L. Artesunate Alleviates Interleukin-1beta-Induced Inflammatory Response and Apoptosis by Inhibiting the NF-kappaB Signaling Pathway in Chondrocyte-Like ATDC5 Cells, and Delays the Progression of Osteoarthritis in a Mouse Model. Int. J. Mol. Med. 2019, 44, 1541–1551. [CrossRef]
[19] Zhao, C.; Zhao, L.; Zhou, Y.; Feng, Y.; Li, N.; Wang, K. Artesunate Ameliorates Osteoarthritis Cartilage Damage by Updating MTA1 Expression and Promoting the Transcriptional Activation of LXA4 to Suppress the JAK2/STAT3 Signaling Pathway. Hum. Mol. Genet. 2023, 32, 1324–1333. [CrossRef] [PubMed]
[20] Ma, L.; Zhao, X.; Liu, Y.; Wu, J.; Yang, X.; Jin, Q. Dihydroartemisinin Attenuates Osteoarthritis by Inhibiting Abnormal Bone Remodeling and Angiogenesis in Subchondral Bone. Int. J. Mol. Med. 2021, 47, 04855. [CrossRef]
[21] Baell, J.B.; Holloway, G.A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. [CrossRef] [PubMed]
[22] Xue, S.T.; Li, K.; Gao, Y.; Zhao, L.Y.; Gao, Y.; Yi, H.; Jiang, J.D.; Li, Z.R. The Role of the Key Autophagy Kinase ULK1 in Hepatocellular Carcinoma and Its Validation as a Treatment Target. Autophagy 2020, 16, 1823–1837. [CrossRef] [PubMed]
[23] Neog, N.; Puzari, M.; Chetia, P. Identification of Potential Inhibitors of Three NDM Variants of Klebsiella Species from Natural Compounds: A Molecular Docking, Molecular Dynamics Simulation, and MM-PBSA Study. Curr. Comput. Aided Drug Des. 2025, 21, 142–165. [CrossRef] [PubMed]
[24] Alotaibi, B.S. In Silico Identification of Phytochemical Inhibitors for Multidrug-Resistant Tuberculosis Based on Novel Pharmacophore Generation and Molecular Dynamics Simulation Studies. BMC Chem. 2024, 18. [PubMed] [CrossRef]
[25] Li, Y.; Li, L.; Wang, S.; Tang, X. EQUIBIND: A Geometric Deep Learning-Based Protein-Ligand Binding Prediction Method. Drug Discov. Ther. 2023, 17, 363–364. [CrossRef] [PubMed]
[26] Lu, W.; Wu, Q.; Zhang, J.; Rao, J.; Li, C.; Zheng, S.J. Tankbind: Trigonometry-Aware Neural Networks for Drug-Protein Binding Structure Prediction. bioRxiv 2022, 35, 7236–7249. [CrossRef]
[27] Xia, S.; Gu, Y.; Zhang, Y. Normalized Protein-Ligand Distance Likelihood Score for End-to-End Blind Docking and Virtual Screening. J. Chem. Inf. Model. 2025, 65, 1101–1114. [CrossRef]
[28] Xue, M.; Liu, B.; Cao, S.; Huang, X.J.n.D.D. FeatureDock for Protein-Ligand Docking Guided by Physicochemical Feature-Based Local Environment Learning Using Transformer. npj Drug Discov. 2025, 2, 4. [CrossRef]
[29] Wang, Y.; Li, J.; Li, X.; Gao, B.; Chen, J.; Song, Y. Spectroscopic and Molecular Docking Studies on Binding Interactions of Camptothecin Drugs with Bovine Serum Albumin. Sci. Rep. 2025, 15. [CrossRef]
[30] Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [CrossRef]
[31] Seeliger, D.; de Groot, B.L. Ligand Docking and Binding Site Analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010, 24, 417–422. [CrossRef] [PubMed]
[32] Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated Docking with Selective Receptor Flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [PubMed] [CrossRef]
[33] Lima, L.R.; Bastos, R.S.; Ferreira, E.F.B.; Leao, R.P.; Araujo, P.H.F.; Pita, S.; De Freitas, H.F.; Espejo-Roman, J.M.; Dos Santos, E.; Ramos, R.D.S.; et al. Identification of Potential New Aedes Aegypti Juvenile Hormone Inhibitors from N-Acyl Piperidine Derivatives: A Bioinformatics Approach. Int. J. Mol. Sci. 2022, 23. [CrossRef] [PubMed]
[34] Govindharaj, D.; Nachimuthu, S.; Gonsalves, D.F.; Kothandan, R.; Dhurai, B.; Rajamani, L.; Ramakrishana, S. Molecular Docking Analysis of Chlorogenic Acid Against Matrix Metalloproteinases (MMPs). Biointerface Res. Appl. Chem. 2020, 10, 6865–6873. [CrossRef]
[35] Gimeno, A.; Cuffaro, D.; Nuti, E.; Ojeda-Montes, M.J.; Beltran-Debon, R.; Mulero, M.; Rossello, A.; Pujadas, G.; Garcia-Vallve, S. Identification of Broad-Spectrum MMP Inhibitors by Virtual Screening. Molecules 2021, 26. [CrossRef]
[36] O’Brien, P.M.; Ortwine, D.F.; Pavlovsky, A.G.; Picard, J.A.; Sliskovic, D.R.; Roth, B.D.; Dyer, R.D.; Johnson, L.L.; Man, C.F.; Hallak, H. Structure-Activity Relationships and Pharmacokinetic Analysis for a Series of Potent, Systemically Available Biphenylsulfonamide Matrix Metalloproteinase Inhibitors. J. Med. Chem. 2000, 43, 156–166. [CrossRef] [PubMed]
[37] Szczygielski, O.; Dabrowska, E.; Niemyjska, S.; Przylipiak, A.; Zajkowska, M. Targeting Matrix Metalloproteinases and Their Inhibitors in Melanoma. Int. J. Mol. Sci. 2024, 25. [CrossRef] [PubMed]
[38] Ding, Y.; Li, Q.; Tang, W.; Pan, Y.; Nie, R.; Meng, X. Effect of a Primer Containing Artemisinin on Dentin Endogenous MMPs and Resin Bond Durability. Int. J. Adhes. Adhes. 2023, 123, 103340. [CrossRef]
[39] Choi, J.Y.; Chung, E. Molecular Dynamics Simulations of Matrix Metalloproteinase 13 and the Analysis of the Specificity Loop and the S1’-Site. Int. J. Mol. Sci. 2023, 24. [CrossRef]
[40] DeJulius, C.R.; Walton, B.L.; Colazo, J.M.; d’Arcy, R.; Francini, N.; Brunger, J.M.; Duvall, C.L. Engineering Approaches for RNA-Based and Cell-Based Osteoarthritis Therapies. Nat. Rev. Rheumatol. 2024, 20, 81–100. [CrossRef] [PubMed]
[41] Paesa, M.; Alejo, T.; Garcia-Alvarez, F.; Arruebo, M.; Mendoza, G. New Insights in Osteoarthritis Diagnosis and Treatment: Nano-Strategies for an Improved Disease Management. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15. [CrossRef] [PubMed]
[42] Xu, H.W.; Li, W.F.; Hong, S.S.; Shao, J.J.; Chen, J.H.; Chattipakorn, N.; Wu, D.; Luo, W.; Liang, G. Tabersonine, a Natural NLRP3 Inhibitor, Suppresses Inflammasome Activation in Macrophages and Attenuate NLRP3-Driven Diseases in Mice. Acta Pharmacol. Sin. 2023, 44, 1252–1261. [CrossRef] [PubMed]
[43] Wang, S.; Shi, X.; Li, J.; Huang, Q.; Ji, Q.; Yao, Y.; Wang, T.; Liu, L.; Ye, M.; Deng, Y.; et al. A Small Molecule Selected from a DNA-Encoded Library of Natural Products That Binds to TNF-alpha and Attenuates Inflammation In Vivo. Adv. Sci. (Weinh.) 2022, 9, e2201258. [CrossRef] [PubMed]
[44] Wei, Y.; Bai, L. Recent Advances in the Understanding of Molecular Mechanisms of Cartilage Degeneration, Synovitis and Subchondral Bone Changes in Osteoarthritis. Connect. Tissue Res. 2016, 57, 245–261. [CrossRef]
[45] Rowan, A.D.; Litherland, G.J.; Hui, W.; Milner, J.M. Metalloproteases as Potential Therapeutic Targets in Arthritis Treatment. Expert. Opin. Ther. Targets 2008, 12, 1–18. [CrossRef]
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more