
APA Style
Abdo Meyiah, Mohammad A. Al-Mterin, Khaled Murshed, Eyad Elkord. (2025). Association of Expression of Immune Checkpoints in Tumor-Infiltrating CD4+ T Lymphocytes with Disease-Free Survival in Colorectal Cancer Patients. Cancer Immunology Connect, 1 (Article ID: 0006). https://doi.org/10.69709/CIConnect.2025.119991MLA Style
Abdo Meyiah, Mohammad A. Al-Mterin, Khaled Murshed, Eyad Elkord. "Association of Expression of Immune Checkpoints in Tumor-Infiltrating CD4+ T Lymphocytes with Disease-Free Survival in Colorectal Cancer Patients". Cancer Immunology Connect, vol. 1, 2025, Article ID: 0006, https://doi.org/10.69709/CIConnect.2025.119991.Chicago Style
Abdo Meyiah, Mohammad A. Al-Mterin, Khaled Murshed, Eyad Elkord. 2025. "Association of Expression of Immune Checkpoints in Tumor-Infiltrating CD4+ T Lymphocytes with Disease-Free Survival in Colorectal Cancer Patients." Cancer Immunology Connect 1 (2025): 0006. https://doi.org/10.69709/CIConnect.2025.119991.
 ACCESS
Research Article
ACCESS
Research Article
Volume 1, Article ID: 2025.0006

Abdo Meyiah
Abdo.Meyiah@xjtlu.edu.cn

Mohammad A. Al-Mterin
mhm8160506@ju.edu.jo

Khaled Murshed
kmurshed@hamad.qa

Eyad Elkord
eyad.elkord@xjtlu.edu.cn
1 Department of Biosciences and Bioinformatics & Suzhou Municipal Key Lab of Biomedical Sciences and Translational Immunology, School of Science, Xi’an Jiaotong-Liverpool University, Suzhou 215123, Jiangsu, China
2 Department of Pharmacology, School of Medicine, The University of Jordan, Amman, Jordan
3 Department of Pathology, Hamad Medical Corporation, Doha, Qatar
4 College of Health Sciences, Abu Dhabi University, Abu Dhabi, United Arab Emirates
5 Biomedical Research Center, School of Science, Engineering and Environment, University of Salford, Manchester, UK
* Author to whom correspondence should be addressed
Received: 12 Feb 2025 Accepted: 18 Mar 2025 Available Online: 20 Mar 2025 Published: 15 Apr 2025
Objectives: The role of inhibitory immune checkpoints (ICs), which are expressed on tumor-infiltrating T lymphocytes (TILs) in colorectal cancer (CRC), and their associations with disease-free survival (DFS) have not been fully elucidated. Methods: This study evaluated the associations between ICs expressed on CD4+ TILs and DFS in forty-five treatment-naïve CRC patients. Initially, we investigated the associations of ICs, including PD-1, TIGIT, LAG-3, and TIM-3, expressed on CD4+ TILs with DFS. Results: There were no statistically significant differences in the associations between the levels of all single IC-expressing CD4+ TILs and DFS. Interestingly, when PD-1 was combined with other ICs, we found that a lack of PD-1 expression on LAG-3+CD4+ TILs (PD-1−LAG-3+) was significantly associated with shorter DFS. Additionally, a lack of expression of both PD-1 and TIGIT (PD-1−TIGIT−) on CD4+ TILs was associated with shorter DFS. Conclusion: We identified two CD4+ T-cell subpopulations (PD-1−LAG-3+ and PD-1−TIGIT−) associated with worse DFS. Our findings highlight the importance of investigating multiple IC co-expressions to determine the exact subpopulations associated with patient prognoses. This preliminary study requires further validation in a larger cohort of CRC patients.
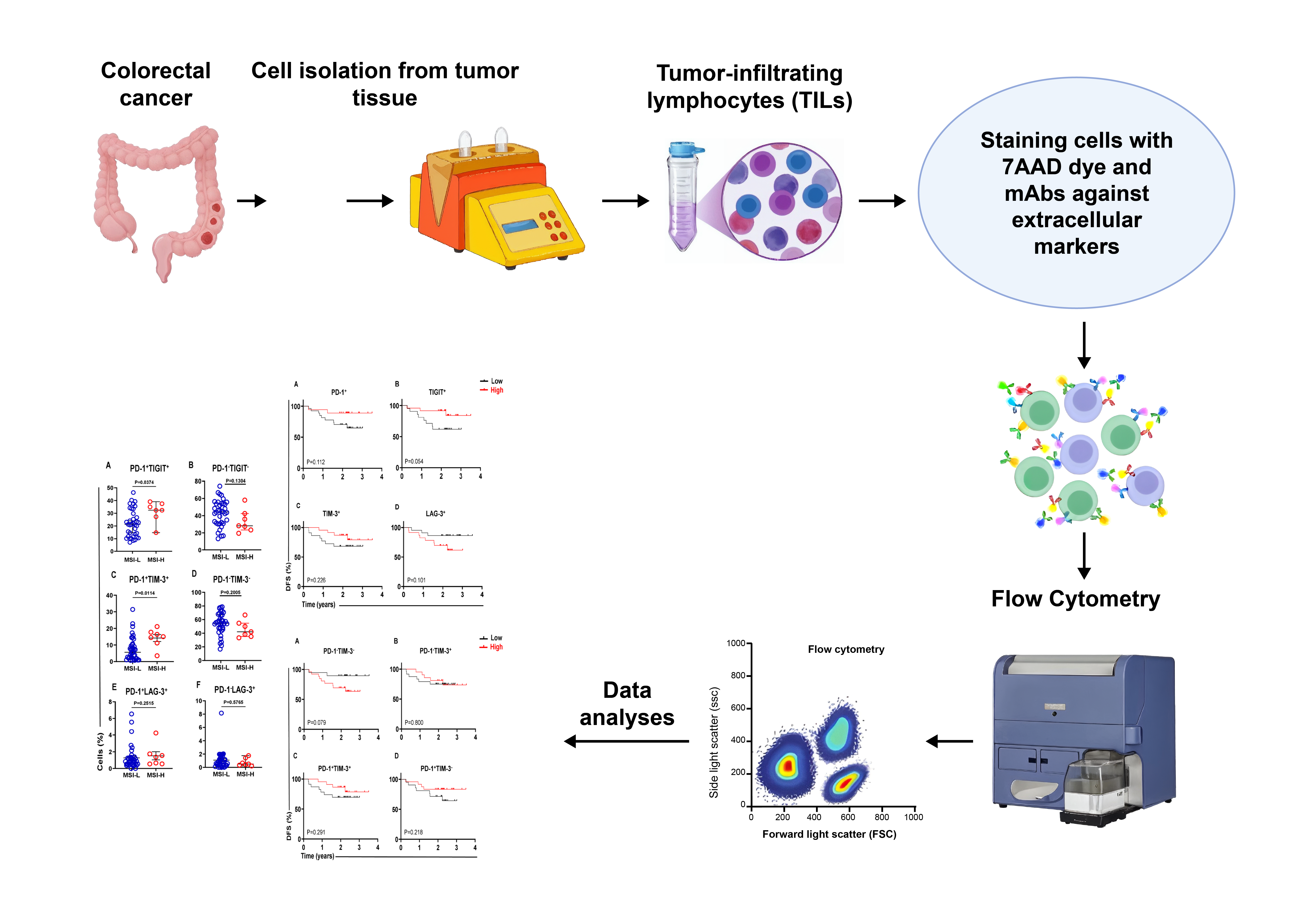
Globally, colorectal cancer (CRC) is the third most common cancer among men and women, and the second most fatal tumor after lung cancer [1]. Many CRC cases are diagnosed at advanced stages as a result of the absence of clinical symptoms during the disease’s early stages, which contributes to poor prognoses and reduced survival rates, in addition to complicating treatment options. Microsatellite instability (MSI) is reported in 10–15% of sporadic CRC and is generally caused by genetic changes/mutations in DNA mismatch repair (MMR) genes. CRC patients with high microsatellite instability/MMR-deficient (MSI-high/dMMR) status have a good prognosis and longer survival rates than patients with microsatellite stable/microsatellite instability-low (MSS/MSI-Low) status [2,3,4]. Within the complex battlefield of the tumor microenvironment (TME), immune cells have the potential to recognize and kill cancer cells, playing key roles in tumor regression and the response to different treatment modalities. Tumor-infiltrating lymphocytes (TILs) are important components of the TME and represent specific histological features of different tumors. TILs consist of varied immune cells such as CD8+ and CD4+ T cells, T regulatory cells (Tregs), and other immune cells. In general, the presence of TILs is associated with increased survival rates and improved response to different treatment modalities in CRC patients [5,6]. However, CD4+ T-cell subsets can either promote or suppress the antitumor CD8+ cytotoxic T-cell responses [7]. The importance of CD4+ T lymphocytes in developing and maintaining an effective antitumor immune response has recently become more evident, even in the context of biological therapies such as cancer immunotherapy that target and activate antitumor immune responses of CD8+ CTLs [8]. Some studies have reported that high-density CD4+ TILs are correlated with improved overall survival (OS) and disease-free survival (DFS) in different types of cancers [9,10,11,12]. Immune checkpoint (IC) molecules and their ligands are highly expressed in the TME. Interactions between ICs and their ligands are critical for inhibiting antitumor immune responses, and blocking these interactions has been shown to improve antitumor immune and clinical responses in different cancers. ICs, including T-cell immunoreceptor with Ig and ITIM domains (TIGIT), programmed cell death-1 (PD-1, CD279), lymphocyte activation gene-3 (LAG-3), T-cell immunoglobulin and mucin domain-3 (TIM-3), and others, play essential roles in immune responses through restraining T lymphocyte activation pathways [13]. High expression of ICs on CD4+ T cells can act as a mechanism for tumor promotion and immune evasion, particularly by inhibiting the function of tumor-specific T cells. For instance, PD-1 expressed on T cells binds to PD-L1 expressed on tumor cells and antigen-presenting cells, leading to functional exhaustion of T cells and impairment of anti-tumor immune responses. Therefore, understanding the role of ICs in regulating the function of TILs should provide insights into novel therapeutic strategies to improve anti-tumor immune responses and enhance the clinical outcomes of cancer patients [14]. This study aimed to evaluate the potential association between the levels of CD4+ TILs expressing different ICs and DFS in treatment-naïve patients with CRC, which might provide potential predictive biomarkers and therapeutic targets for CRC patients.
2.1. Patients and Samples This study was executed under ethical approval from the Institutional Review Board (IRB) of the Medical Research Center at Hamad Medical Corporation (HMC), Doha, Qatar (approval number MRC-02-18-012). Each patient included in this study provided written consent before any sample was collected. Based on the pathologist’s report, fifty tumor tissue (TT) samples were collected from treatment-naïve CRC patients with different tumor-node-metastasis stages (TNM stages I to IV). The collection period ranged from late 2018 to early 2020, and disease progression data were collected in March 2022 for these patients. No patients in this study were lost to follow-up. All treatment-naïve patients with CRC included in this study underwent surgery without any chemo- or radiotherapy before the surgery and sampling. A total of 45 eligible CRC patients were included in the DFS analyses. Details of the CRC patients included in this study are presented in Table 1, which was taken from Meyiah et al. [15]. CRC patients’ clinical and pathological features in this study. 2.2. Cell Isolation from Colon Tumor Tissues A mechanical disaggregation method was used to isolate single-cell suspensions from TT specimens, as we have previously described [16]. In brief, all colon tumor tissues were rinsed in PBS and cut into small pieces. The tissues were subsequently dissociated into single-cell suspensions using a gentlyMACS dissociator (Miltenyi Biotec, Bergisch Gladbach, Germany). Then, the cell suspension was prepared by slowly passing through a sterile cell strainer (size 100 µM) to eliminate any large particles or cell aggregates. The cells were then washed at least 3 times with PBS followed by cell staining for cytometric analyses. 2.3. Staining and Flow Cytometric Analyses The cells were stained with different fluorochrome-conjugated monoclonal antibodies (mAbs), and FACS analysis was performed as described in the previous study [17]. Briefly, the cell suspension was washed once with cold PBS and then resuspended in FACS staining buffer consisting of PBS, 1% FCS, and 0.1% sodium azide, before being blocked with Fc receptor (FcR) blocking reagent (Miltenyi Biotec). 7-Aminoactinomycin D (7-AAD; BioLegend, San Diego, CA, USA) and different mAbs were then added to stain the cells. These mAbs included anti-CD4-phycoerythrin (BD Biosciences, Oxford, UK), anti-CD3-Alexa Fluor 700 (BD Biosciences), anti-LAG-3-BUV421 (BD Biosciences), anti-TIM-3-BV711 (BD Biosciences), anti-PD-1-PE/DazzleTM 594 (BioLegend, San Diego, USA), and anti-TIGIT-APC (eBioscience, San Diego, USA). Following staining, the cells were washed once with ice-cold FACS staining buffer, and centrifuged for 5 min at 400 × g. The supernatant was removed, and the cells were re-suspended in FACS buffer for flow cytometric analyses. 7-ADD dye was used to exclude dead cells and gate only live cells. Fluorescence Minus One (FMO) control was used to define the proper gating strategy. Cells were gated based on the forward scatter area (FSC-A) and the side scatter area (SSC-A), and then lymphocyte gate was applied to select single cells (singlets) based on the FSC-A and the FSC-H. The singlet cell gate was applied to select 7-ADD negative live cells. Live cells were further gated to select CD4+ T cells, and the expression levels of single ICs, including PD-1, TIGIT, TIM-3, and LAG-3 on CD4+ T cells and co-expression of ICs were determined. A BD Biosciences LSRFortessa X-20 flow cytometer and BD FACSDiva® software were used to analyze the samples. FlowJo software (version 10, FlowJo LLC, Ashland, OR, USA) was used to analyze the collected data. All flow cytometric plots used in this study have already been published (please see Figure 3 Toor et al. [17]). 2.4. Statistical Analyses Statistical analyses for all the datasets were performed by using GraphPad Prism version 9 software (GraphPad Prism® Software, California, CA, USA). Parameters, including the mean and median values with their standard error of the mean (SEM), as well as the minimum and maximum values, were calculated using descriptive statistical methods. The normality test was performed using the Shapiro-Wilk test. The cell subsets were classified as low/high if they were below/above the median for nonnormally distributed data, and below/above the mean for normally distributed data. Kaplan- Meier survival curves were generated to estimate and compare DFS between the high and low groups, and the log-rank test was used to calculate the P-values of the survival curves. An unpaired t-test was performed to compare the values between two groups (MSI-high and MSI-low). Statistical significance was defined as a p value ≤ 0.05.
Feature
CRC Patients
Number
45
Median age (range)
56 (18-79)
Gender: Male/Female
30/15
TNM stage
Stage-I
(4 patients)
Stage-II
(20 patients)
Stage-III
(14 patients)
Stage-IV
(7 patients)
Histological grading of tumor
Grade 2 (Moderately tumor differentiated)
(41 patients)
Grade 3 (Poorly tumor differentiated)
(4 patients)
MSI-H and DMMR
(8 patients)
Loss of nuclear expression for MLH1 and PMS2
(7 patients)
Loss of nuclear expression of MSH2
(1 patient)
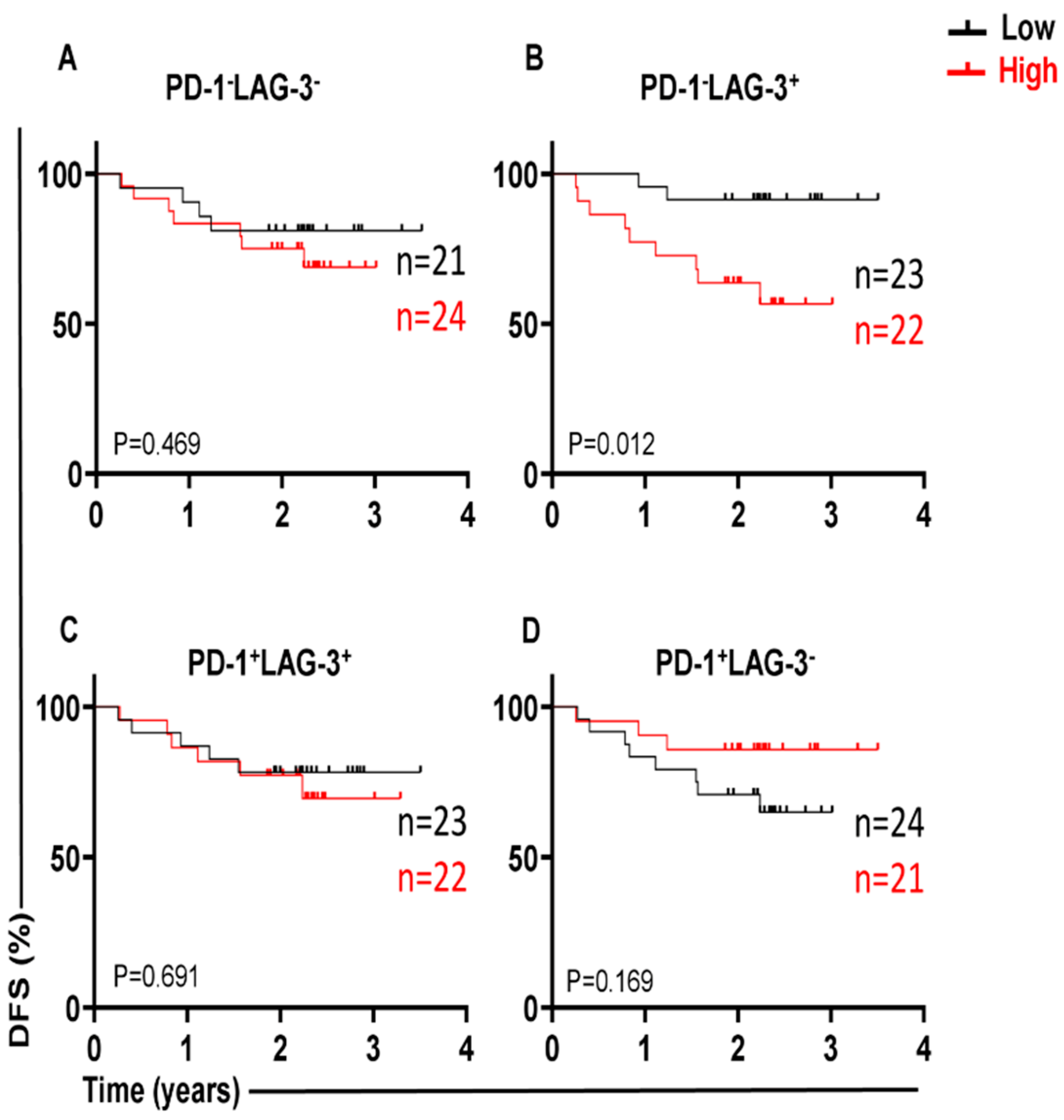
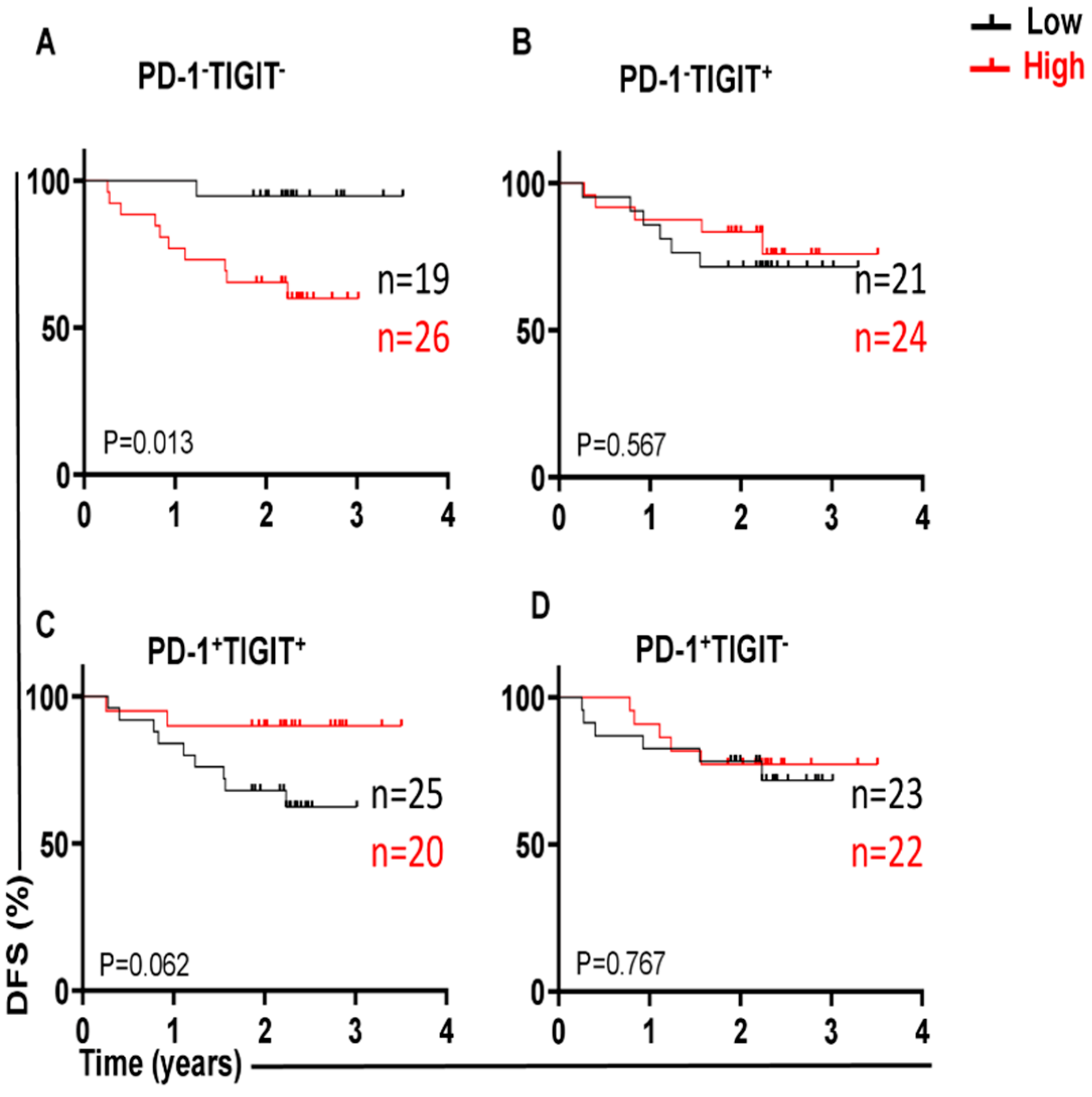
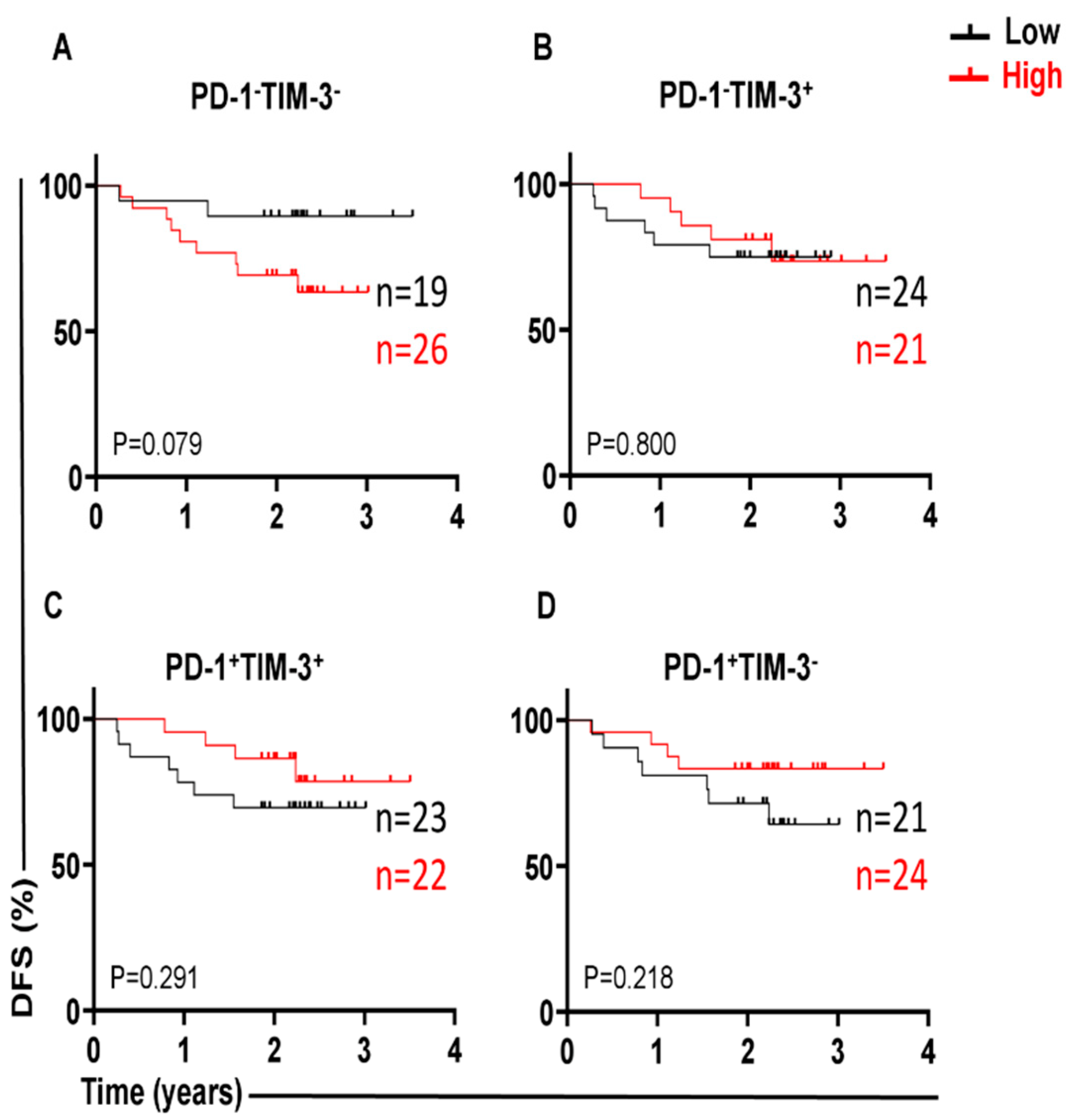
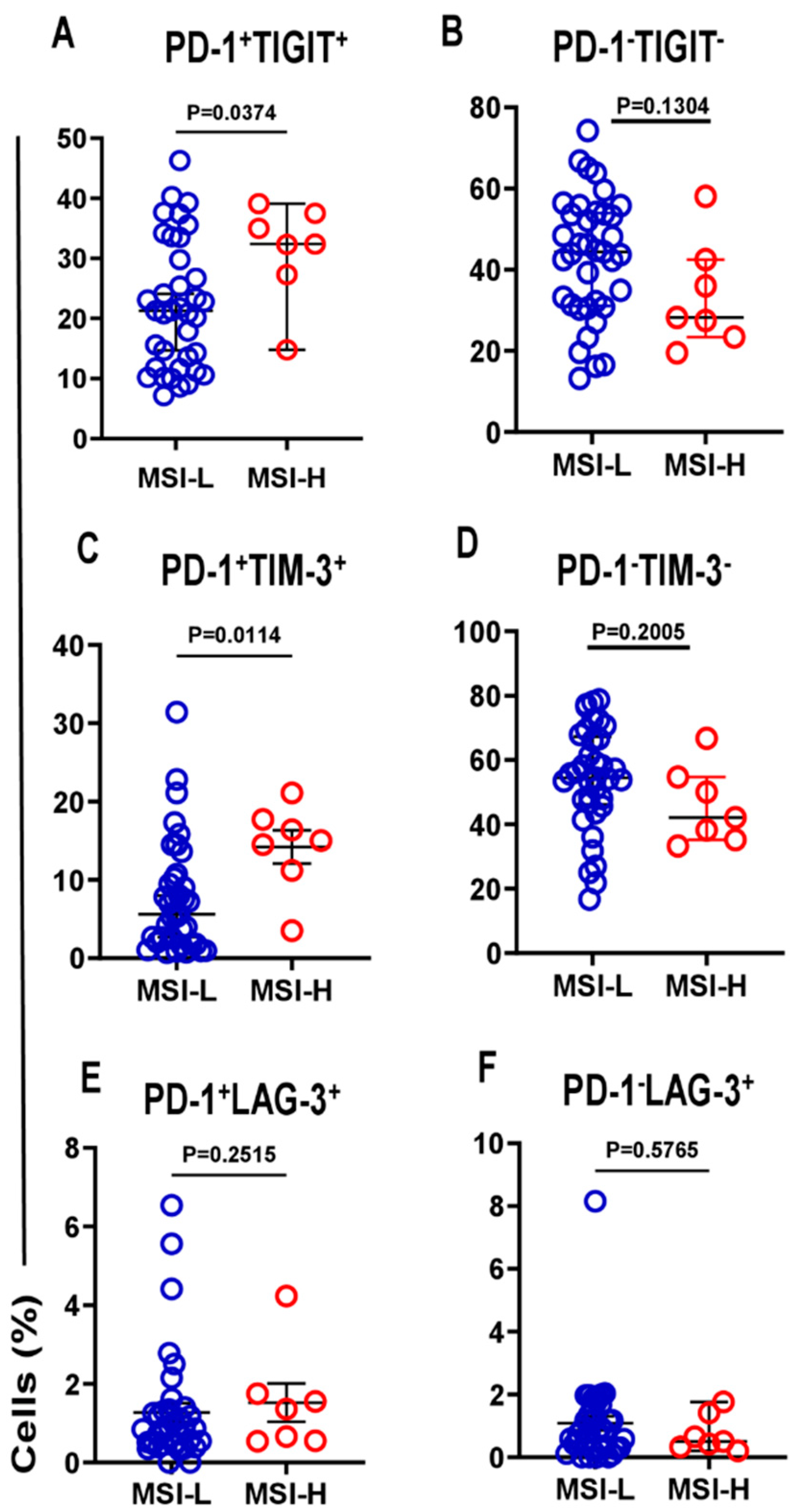
3.1. Different Immune Checkpoint-Expressing CD4 TILs in CRC and Their Associations with DFS Our previous study revealed that CD4+ T-cell subsets are increased in the TME of CRC, and they express several IC molecules, including PD-1, CTLA-4, TIM-3, and LAG-3, playing crucial roles in CRC progression [16]. In the present study, we expanded our previous work to a larger cohort of CRC patients. CD4+ T-cell levels were divided into low and high groups based on the median, and there was no significant difference in terms of DFS between the two groups. Additionally, we compared the percentages of CD4+ T cells between early-stage and advanced-stage CRC patients, and there were no significant differences between them, as shown in Table 2. PD-1 (mean 44.7), TIGIT (mean 36.8), and TIM-3 (median 9.5) were expressed at high levels, whereas LAG-3 (median 1.8) was expressed at low levels on CD4+ TILs. Patients were then categorized into groups above or below the mean/median based on a single IC expression level on CD4+ TILs. Notably, there were no significant differences in terms of DFS between CRC patients with higher percentages (above the mean/median) of tumor-infiltrating CD4+PD-1+ (p = 0.112, Figure 1A), CD4+TIGIT+ (p = 0.054, Figure 1B), CD4+TIM-3+ (p = 0.226, Figure 1C), and CD4+LAG-3+ TILs (p = 0.101, Figure 1D), and groups of patients with low percentages (below the mean/median) of these cell subsets. Percentages of CD4+ T cells and immune checkpoints in CD4+ T cells in early-stage and advanced-stage CRC patients. 3.2. A Lack of PD-1 Expression on LAG-3CD4 TILs Is Associated with Shorter DFS We observed that the expression level of single ICs was significantly associated with DFS. To gain deeper insights, we then investigated the associations of a combination of ICs co-expressed on CD4+ TILs with DFS. We specifically investigated the co-expression of PD-1 with other ICs on CD4+ TILs, which could be associated with DFS. Among the four cell subsets investigated, PD-1±LAG-3± CD4+ TILs (Figure 2), interestingly, there was a significant association between high PD-1−LAG-3+ percentages and shorter DFS (p = 0.012, Figure 2B). In contrast, high frequencies of other cell subsets, including PD-1−LAG-3−, PD-1+LAG-3+, and PD-1+LAG-3− CD4+ TILs were not associated with DFS (Figure 2A,C,D). These findings show that a lack of PD-1 expression on LAG-3+CD4+ TILs is associated with shorter DFS in CRC patients. 3.3. Lack of Co-Expression of PD-1 with TIGIT Is Associated with Shorter DFS We then investigated the associations of PD-1±TIGIT± and PD-1±TIM-3± levels on CD4+ TILs with DFS (Figure 3 and Figure 4). Interestingly, lack of PD-1 and TIGIT expression on CD4+ TILs was significantly associated with shorter DFS (p = 0.013, Figure 3A). In agreement with these findings, patients with higher levels of the two IC co-expression of PD-1+TIGIT+ on CD4+ TILs had longer DFS, but without a significant difference (p = 0.062, Figure 3C). Other cell subsets including PD-1−TIGIT+ (Figure 3B) and PD-1+TIGIT− (Figure 3D) were not associated with DFS. With regards to PD-1±TIM-3± cell subsets, patients with higher percentages of PD-1−TIM-3− CD4+ TILs tended to have shorter DFS than those with lower percentages of CD4+PD-1−TIM-3− TILs, but without any significant difference (p = 0.079, Figure 4A), while other cell subsets did not show any association with DFS (Figure 4B–D). Our data highlight the significance of different IC co-expressions on tumor-infiltrating CD4+ T cells, while their lack of co-expression could be a negative prognostic marker in treatment-naïve CRC patients. 3.4. Differences in Immune Checkpoint Co-Expression on CD4 TILs in Patients with MSI-High and MSI-Low Tumors We have previously shown that CRC patients with dMMR or MSI-high tumors displayed elevated levels of immune checkpoint-expressing T lymphocytes compared with those with MMR-proficient and MSS/MSI-low tumors [17]. Specifically, we reported that MSI-high CRC patients had significantly higher levels of TIGIT+ and TIM-3+CD4+ TILs, as compared with MSI-low CRC [17]. In this study, we analyzed the differences in the percentages of tumor-infiltrating CD4+PD-1+/− T cells co-expressing other ICs (TIGIT, TIM-3, or LAG-3) between MSI-high and MSI-low CRC. Among the 44 patients with MSI data, only 7 patients (16%) had MSI-high. We noticed that CRC patients with MSI-high status had significantly greater percentages of PD-1+TIGIT+ and PD-1+TIM-3+ CD4+ T cells (Figure 5A and Figure 5C, respectively), as compared to patients with MSI-low status (PD-1+TIGIT+: median; MSI-high 32.4 vs. MSI-low 21.3, p = 0.037; PD-1+TIM-3+: median; MSI-high 15.0 vs. MSI-low 5.6, p = 0.011). In line with these findings, levels of CD4+ TILs lacking co-expression of PD-1 plus TIGIT (Figure 5B) or PD-1 plus TIM-3 (Figure 5D) were lower in MSI-high patients than in MSI-low patients. However, statistical significance was not reached, which is more likely due to the small sample size. In addition, there were no significant differences in PD-1+LAG-3+ (Figure 5E) or PD-1−LAG-3− (Figure 5F) CD4+ T cells between MSI-high and MSI-low patients. These findings revealed an increase in IC co-expression in MSI-high CRC, which can be linked to improved prognoses and an enhanced response to immunotherapies.
Early-Stage
(Mean ± SEM)
(Min–Max)Advanced-Stage
(Mean ± SEM)
(Min–Max)
CD4+ T cells
38.5 ± 2.8
17.0–63.638.8 ± 3.4
14.4–74.1
CD4+ PD-1+ T cells
44.4 ± 2.9
19.9–72.445.2 ± 3.8
20.1–79.9
CD4+ TIGIT+ T cells
39.6 ± 2.7
12.0–60.633.7 ± 2.4
10.6–48.6
CD4+ LAG-3+ T cells
2.4 ± 0.6
0.0–14.72.2 ± 0.3
0.0–6.3
CD4+ TIM-3+ T cells
10.5 ± 1.4
1.1–25.310.8 ± 2.0
0.0–34.7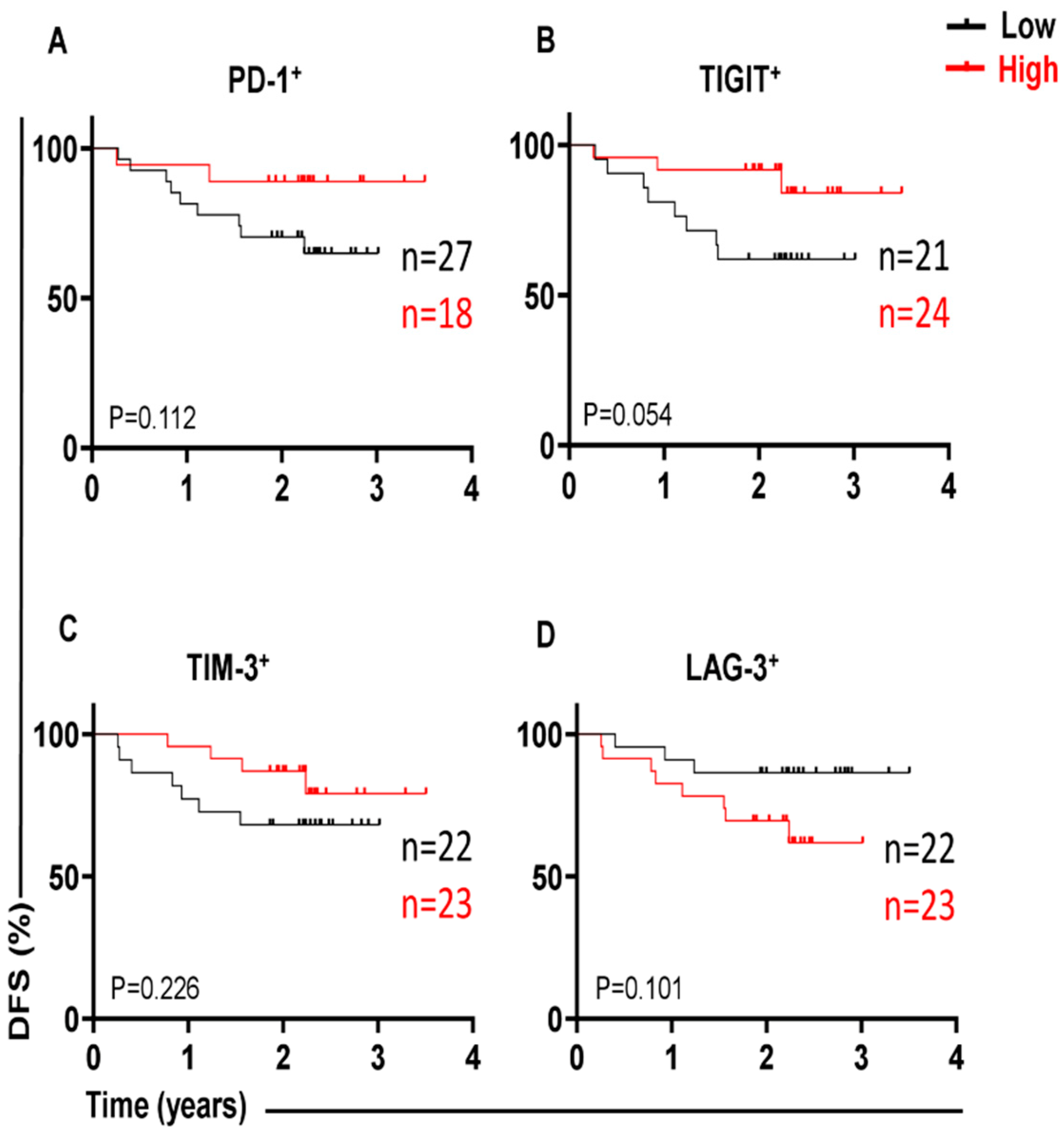
Several studies have documented the prognostic significance of TILs in various malignancies. A high density of TILs in CRC tissues was correlated with a better prognosis and improved survival outcome [18]. CD4+ TILs are essential indicators of antitumor immunity [19], and they have been linked to improved clinical outcomes in various tumor types [20,21], including CRCs [22]. CD4+ T cells in the TME express high numbers of different ICs, including PD-1, LAG-3, TIGIT, and TIM-3 [23]. We have recently shown the associations between different types of IC-expressing CD4+ TILs within tumors and DFS in patients with CRC [24]. Many studies have shown that high expression levels of different ICs on T cells are linked to T-cell dysfunction/exhaustion and prognosis in several types of cancer, including cervical [25], acute myeloid leukemia [26], liver [27], and breast cancers [28]. The purpose of this work was to determine the prognostic significance of IC expression on CD4+ TILs in relation to DFS and MSI status in treatment-naïve patients with CRC. LAG-3 inhibits the proliferation and activation of T-cell subsets through its interaction with a variety of ligands. It has a greater affinity for major histocompatibility complex (MHC) class II (MHC II) signaling than CD4 and might suppress CD4+ T cell activation by inhibiting CD4-MHC II interaction [29,30]. Other studies have demonstrated that the inhibitory function of LAG-3 is not dependent on CD4 competition, but rather relies on its cytoplasmic domain to deliver inhibitory signals [31,32]. Therefore, the expression of LAG-3 in TILs is generally indicative of aggressive progression, and it is linked to worse outcomes in a wide range of human carcinomas [33], including CRC [34,35]. CD4+ TILs display chronic exhaustion traits, followed by the up-regulation of LAG-3 during tumor progression [36]. Thus, it appears to be a promising target for cancer immunotherapies. Some early-stage clinical trials are currently investigating the use of anti-LAG-3 mAbs as therapeutic tools for cancer in a variety of settings [37]. Furthermore, over-expression of LAG-3 in T cells contributes to a reduction in T-cell response [38] and promotes tumor growth in both high/low-grade gliomas. Additionally, higher LAG-3 levels are associated with poorer overall survival prognosis [39]. Studies have reported that overexpression of LAG-3 is associated with poor survival in hepatocellular carcinoma [40], melanoma [41], head and neck squamous cell carcinoma [42], and gastric carcinoma with Epstein-Barr virus (EBV) infection and MLH1 tumor mutations [43]. This overexpression may contribute to tumor growth and progression. Another study showed that blocking LAG-3 may enhance TIL responses in proficient MMR liver metastases of CRC patients, increase the effector cytokine production in the TME, and enhance anti-tumor immune responses [44]. Other studies reported that high levels of LAG-3 in some cancers, including breast [45], non-small cell lung [46], and esophageal squamous cell cancers [47] were associated with improved survival. In this study, shorter DFS was observed in patients with higher percentages of CD4+LAG-3+ TILs, but without showing any statistically significant difference. However, a lack of PD-1 expression on LAG-3+CD4+ TILs was a critical factor, contributing to a significant association with shorter DFS. Our study highlights the significance of the tumor-infiltrating PD-1−LAG-3+CD4+ T-cell subset as a potential prognostic marker in CRC patients. TIGIT is an immunosuppressive receptor, that is expressed mostly on T lymphocytes, including CD4+ T cells. The function of TIGIT has been reported in several cancers, including lung adenocarcinoma [48] and hepatocellular carcinoma [49]; however, its function in CRC remains unclear. We previously showed that high expression of TIGIT in the circulation of CRC patients was associated with good prognoses in the early disease stage [50]. Another study reported that overexpression of TIGIT was associated with improved survival in many cancers [51]. Moreover, the overexpression of PD-1 and TIGIT in tumor tissues with dMMR status was associated with advanced TNM stage and better DFS [52]. In contrast, other studies have shown that the overexpression of TIGIT is associated with CD8+ T-cell exhaustion, advanced disease, and decreased survival rates [53]. In this study, we reported that patients with high percentages of CD4+TIGIT+ T cell infiltration had longer DFS, but this difference was not statistically significant. Similar to our findings, high TIGIT expression levels are associated with good OS in CRC patients [54]. Co-expression of PD-1 and TIGIT is associated with T-cell dysfunction in cancer [55]. The co-expression of multiple types of ICs has been reported in the microenvironment of many cancers. However, few studies have focused on the association between the co-expression of different ICs and cancer patient prognosis. A previous study indicated that PD-L1 co-expression with either TIM-3 or TIGIT reduced the OS rate in esophageal squamous cell carcinoma patients [56]. In advanced gastrointestinal cancer, patients with high frequencies of PD-1+ and TIM-3+ cells among total T-cell subsets exhibited poor clinical outcomes according to multivariate analysis [57]. However, our previous study showed that high expression levels of TIM-3 on CD8+ T cells in both circulation and the TME were associated with longer DFS in CRC patients [58]. Moreover, a recent study reported that a higher frequency of PD-1+TIM-3+ cells in CD4+ and CD8+ T-cell populations could be present in AML patients than in healthy individuals and AML-complete remission groups [59]. Additionally, increased co-expression of PD-1 with TIM-3 was associated with CD8+ T-cell exhaustion and poor prognosis in AML patients [60]. Of note, co-expression of PD-1/TIM-3 on T cells may induce resistance to the immune checkpoint inhibitors in CRC patients [61]. Our study revealed that a lack of co-expression of PD-1 with TIGIT or PD-1 with TIM-3 is associated with poor DFS in CRC patients. Additionally, we found that the levels of PD-1+TIGIT+ and PD-1+TIM-3+ infiltrating CD4+ T cells were greater in the MSI-high, which is associated with a good prognosis. Of note, PD-1 with TIGIT or PD-1 with TIM-3 was investigated mostly in bulk tumors, whereas our study investigated IC co-expressions in T-cell subsets. Examining soluble ICs in the serum of cancer patients could be useful biomarkers for patients’ prognoses. A recent study showed that high levels of soluble PD-L1 can be detected in CRC patients’ serum during tumor progression and metastases [62,63].
In this study, we identified two CD4+ T-cell subsets (PD-1−LAG-3+ and PD-1−TIGIT−), which are associated with worse DFS. Our data highlight the importance of investigating IC co-expression for more accurate identification of critical cell subsets, which are associated with DFS in CRC patients. However, there are some limitations in this study as well. More specifically, the sample size was relatively small, and the data were divided based on mean/median levels, which could generate only initial hypotheses. Alternative statistical methods for optimizing prognostic separation in training and validation sets should provide more robust findings. Further, larger validation cohorts are needed to confirm these findings. Moreover, additional investigations are required to determine the precise functions of these cell subsets in the tumor microenvironment of CRC patients.
| DFS | Disease-Free Survival |
| CRC | Colorectal Cancer |
| IC | Immune Checkpoint |
| LAG-3 | Lymphocyte Activation Gene-3 |
| MSI | Microsatellite Instability |
| MMR | Mismatch Repair |
| MHC | Major Histocompatibility Complex |
| MHC II | Class II |
| OS | Overall Survival |
| PD-1 | Programmed Cell Death-1 |
| TME | Tumor Microenvironment |
| TILs | Tumor-Infiltrating Lymphocytes |
| Tregs | T Regulatory Cells |
| TIGIT | T-Cell Immunoreceptor with Ig and ITIM Domains |
| TIM-3 | T-Cell Immunoglobulin and Mucin Domain-3 |
| TT | Tumor Tissue |
| 7-AAD | 7-Aminoactinomycin D |
E.E. Conceptualization; A.M., M.A.A.-M. and E.E. Data curation; A.M., M.A.A.-M. and E.E. Formal analysis; E.E. Funding acquisition; E.E. and K.M. Investigation; E.E. Methodology; K.M. Sample acquisition; E.E. Supervision; E.E. Project administration; A.M. and E.E. Writing—original draft. All authors have read and agreed to the published version of the manuscript.
The datasets used and/or analyzed in the present study are available from the corresponding author upon reasonable request.
The current study was approved by the Institutional Review Board (IRB) of the Medical Research Center at Hamad Medical Corporation (HMC), Doha, Qatar (the approval number MRC-02-18-012). Each patient included in this study provided written consent before collecting any sample.
This study was conducted according to the guidelines of the Declaration of Helsinki.
Not applicable.
This research received no external funding.
The authors declare no conflicts of interest.
We would like to thank all patients for donating samples.
[1] F. Bray, J. Ferlay, I. Soerjomataram, R.L. Siegel, L.A. Torre, A. Jemal, "Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries" CA Cancer J. Clin., vol. 68, pp. 394-424, 2018. [Crossref]
[2] P. Maby, G. Bindea, B. Mlecnik, J. Galon, "License to Kill: Microsatellite Instability and Immune Contexture" Oncoimmunology, vol. 10, p. 1905935, 2021. [Crossref] [PubMed]
[3] A. Lin, J. Zhang, P. Luo, "Crosstalk Between the MSI Status and Tumor Microenvironment in Colorectal Cancer" Front. Immunol., vol. 11, 2020. [Crossref] [PubMed]
[4] A. Ooki, E. Shinozaki, K. Yamaguchi, "Immunotherapy in Colorectal Cancer: Current and Future Strategies" J. Anus Rectum Colon., vol. 5, pp. 11-24, 2021. [Crossref]
[5] V. Deschoolmeester, M. Baasy, F. Lardon, P. Pauwels, M. Peeters, "Immune Cells in Colorectal Cancer: Prognostic Relevance and Role of MSI" Cancer Microenviron. Off. J. Int. Cancer Microenviron. Soc., vol. 4, pp. 377-392, 2011. [Crossref]
[6] M. Wozniakova, J. Skarda, M. Raska, "The Role of Tumor Microenvironment and Immune Response in Colorectal Cancer Development and Prognosis" Pathol. Oncol. Res. POR, vol. 28, p. 1610502, 2022. [Crossref]
[7] D.S. Kravtsov, A.K. Erbe, P.M. Sondel, A.L. Rakhmilevich, "Roles of CD4+ T Cells as Mediators of Antitumor Immunity" Front. Immunol., vol. 13, 2022. [Crossref] [PubMed]
[8] R.E. Tay, E.K. Richardson, H.C. Toh, "Revisiting the Role of CD4+ T Cells in Cancer Immunotherapy—New Insights into Old Paradigms" Cancer Gene Ther., vol. 28, pp. 5-17, 2021. [Crossref]
[9] M.P. Pinto, C. Balmaceda, M.L. Bravo, S. Kato, A. Villarroel, G.I. Owen, et al., "Patient Inflammatory Status and CD4+/CD8+ Intraepithelial Tumor Lymphocyte Infiltration Are Predictors of Outcomes in High-Grade Serous Ovarian Cancer" Gynecol. Oncol., vol. 151, pp. 10-17, 2018. [Crossref]
[10] R. Hoesli, A.C. Birkeland, A.J. Rosko, M. Issa, K.L. Chow, N.L. Michmerhuizen, et al., "Proportion of CD4 and CD8 Tumor Infiltrating Lymphocytes Predicts Survival in Persistent/Recurrent Laryngeal Squamous Cell Carcinoma" Oral. Oncol., vol. 77, pp. 83-89, 2018. [Crossref]
[11] D. Borsetto, M. Tomasoni, K. Payne, J. Polesel, A. Deganello, P. Bossi, et al., "Prognostic Significance of CD4+ and CD8+ Tumor-Infiltrating Lymphocytes in Head and Neck Squamous Cell Carcinoma: A Meta-Analysis" Cancers, vol. 13, 2021. [Crossref]
[12] J. Palomero, C. Panisello, M. Lozano-Rabella, R. Tirtakasuma, J. Díaz-Gómez, D. Grases, et al., "Biomarkers of Tumor-Reactive CD4(+) and CD8(+) TILs Associate with Improved Prognosis in Endometrial Cancer" J. Immunother. Cancer, vol. 10, p. e005443, 2022. [Crossref] [PubMed]
[13] D.M. Pardoll, "The Blockade of Immune Checkpoints in Cancer Immunotherapy" Nat. Rev. Cancer, vol. 12, pp. 252-264, 2012. [Crossref] [PubMed]
[14] B.H. Alsaafeen, B.R. Ali, E. Elkord, "Resistance Mechanisms to Immune Checkpoint Inhibitors: Updated Insights" Mol. Cancer, vol. 24, p. 20, 2025. [Crossref]
[15] A. Meyiah, G. Mahmoodi Chalbatani, M.A. Al-Mterin, M.A. Malekraeisi, K. Murshed, E. Elkord, "Co-Expression of PD-1 with TIGIT or PD-1 with TIM-3 on Tumor-Infiltrating CD8(+) T Cells Showed Synergistic Effects on Improved Disease-Free Survival in Treatment-Naïve CRC Patients" Int. Immunopharmacol., vol. 119, p. 110207, 2023. [Crossref]
[16] S.M. Toor, K. Murshed, M. Al-Dhaheri, M. Khawar, M. Abu Nada, E. Elkord, "Immune Checkpoints in Circulating and Tumor-Infiltrating CD4(+) T Cell Subsets in Colorectal Cancer Patients" Front. Immunol., vol. 10, 2019. [Crossref]
[17] S.M. Toor, V. Sasidharan Nair, K. Murshed, M. Abu Nada, E. Elkord, "Tumor-Infiltrating Lymphoid Cells in Colorectal Cancer Patients with Varying Disease Stages and Microsatellite Instability-High/Stable Tumors" Vaccines, vol. 9, 2021. [Crossref]
[18] A.M. Dahlin, M.L. Henriksson, B. Van Guelpen, R. Stenling, A. Oberg, J. Rutegård, et al., "Colorectal Cancer Prognosis Depends on T-Cell Infiltration and Molecular Characteristics of the Tumor" Mod. Pathol., vol. 24, pp. 671-682, 2011. [Crossref]
[19] M. Ben Khelil, Y. Godet, S. Abdeljaoued, C. Borg, O. Adotévi, R. Loyon, "Harnessing Antitumor CD4(+) T Cells for Cancer Immunotherapy" Cancers, vol. 14, 2022. [Crossref]
[20] J. Galon, A. Costes, F. Sanchez-Cabo, A. Kirilovsky, B. Mlecnik, C. Lagorce-Pagès, et al., "Type, Density, and Location of Immune Cells Within Human Colorectal Tumors Predict Clinical Outcome" Science, vol. 313, pp. 1960-1964, 2006. [Crossref]
[21] B. Mlecnik, M. Tosolini, A. Kirilovsky, A. Berger, G. Bindea, T. Meatchi, et al., "Histopathologic-Based Prognostic Factors of Colorectal Cancers Are Associated with the State of the Local Immune Reaction" J. Clin. Oncol., vol. 29, pp. 610-618, 2011. [Crossref]
[22] I. Atreya, M.F. Neurath, "Immune Cells in Colorectal Cancer: Prognostic Relevance and Therapeutic Strategies" Expert. Rev. Anticancer. Ther., vol. 8, pp. 561-572, 2008. [Crossref]
[23] R. Fromentin, W. Bakeman, M.B. Lawani, G. Khoury, W. Hartogensis, S. DaFonseca, et al., "CD4+ T Cells Expressing PD-1, TIGIT and LAG-3 Contribute to HIV Persistence During ART" PLoS Pathog., vol. 12, 2016. [Crossref]
[24] M.A. Al-Mterin, K. Murshed, A. Alsalman, A. Abu-Dayeh, E. Elkord, "Associations of Different Immune Checkpoints-Expressing CD4+ Treg/ T Cell Subsets with Disease-Free Survival in Colorectal Cancer Patients" BMC Cancer, vol. 22, 2022. [Crossref]
[25] Y. Wang, S. Zhao, X. Zhang, H. Zhu, X. Ji, Y. Jiang, et al., "Higher T cell Immunoglobulin Mucin-3 (Tim-3) Expression in Cervical Cancer Is Associated with a Satisfactory Prognosis" Transl. Cancer Res., vol. 9, pp. 2801-2813, 2020. [Crossref]
[26] Y. Kong, L. Zhu, T.D. Schell, J. Zhang, D.F. Claxton, W.C. Ehmann, et al., "T-Cell Immunoglobulin and ITIM Domain (TIGIT) Associates with CD8+ T-Cell Exhaustion and Poor Clinical Outcome in AML Patients" Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res., vol. 22, pp. 3057-3066, 2016. [Crossref]
[27] D. Ostroumov, S. Duong, J. Wingerath, N. Woller, M.P. Manns, K. Timrott, et al., "Transcriptome Profiling Identifies TIGIT as a Marker of T-Cell Exhaustion in Liver Cancer" Hepatology, vol. 73, pp. 1399-1418, 2021. [Crossref] [PubMed]
[28] S. Muenst, S.D. Soysal, F. Gao, E.C. Obermann, D. Oertli, W.E. Gillanders, "The Presence of Programmed Death 1 (PD-1)-Positive Tumor-Infiltrating Lymphocytes Is Associated with Poor Prognosis in Human Breast Cancer" Breast Cancer Res. Treat., vol. 139, pp. 667-676, 2013. [Crossref]
[29] C.J. Workman, D.S. Rice, K.J. Dugger, C. Kurschner, D.A. Vignali, "Phenotypic Analysis of the Murine CD4-Related Glycoprotein, CD223 (LAG-3)" Eur. J. Immunol., vol. 32, pp. 2255-2263, 2002. [Crossref]
[30] B. Huard, P. Prigent, M. Tournier, D. Bruniquel, F. Triebel, "CD4/Major Histocompatibility Complex Class II Interaction Analyzed with CD4- and Lymphocyte Activation Gene-3 (LAG-3)-Ig Fusion Proteins" Eur. J. Immunol., vol. 25, pp. 2718-2721, 1995. [Crossref]
[31] L.P. Andrews, A.E. Marciscano, C.G. Drake, D.A. Vignali, "LAG3 (CD223) as a Cancer Immunotherapy Target" Immunol. Rev., vol. 276, pp. 80-96, 2017. [Crossref]
[32] T. Maruhashi, I.M. Okazaki, D. Sugiura, S. Takahashi, T.K. Maeda, K. Shimizu, et al., "LAG-3 Inhibits the Activation of CD4(+) T Cells that Recognize Stable pMHCII Through Its Conformation-Dependent Recognition of pMHCII" Nat. Immunol., vol. 19, pp. 1415-1426, 2018. [Crossref]
[33] S. Moerdler, M. Ewart, D.L. Friedman, K. Kelly, Q. Pei, M. Peng, et al., "LAG-3 Is Expressed on a Majority of Tumor Infiltrating Lymphocytes in Pediatric Hodgkin Lymphoma" Leuk. Lymphoma, vol. 62, pp. 606-613, 2021. [Crossref]
[34] N. Sauer, W. Szlasa, L. Jonderko, M. Oślizło, D. Kunachowicz, J. Kulbacka, et al., "LAG-3 as a Potent Target for Novel Anticancer Therapies of a Wide Range of Tumors" Int. J. Mol. Sci., vol. 23, 2022. [Crossref]
[35] J. Chen, Z. Chen, "The Effect of Immune Microenvironment on the Progression and Prognosis of Colorectal Cancer" Med. Oncol., vol. 31, p. 82, 2014. [Crossref]
[36] S.R. Goding, K.A. Wilson, Y. Xie, K.M. Harris, A. Baxi, A. Akpinarli, et al., "Restoring Immune Function of Tumor-Specific CD4+ T Cells During Recurrence of Melanoma" J. Immunol., vol. 190, pp. 4899-4909, 2013. [Crossref]
[37] J.L. Huo, Y.T. Wang, W.J. Fu, N. Lu, Z.S. Liu, "The Promising Immune Checkpoint LAG-3 in Cancer Immunotherapy: From Basic Research to Clinical Application" Front. Immunol., vol. 13, 2022. [Crossref]
[38] X. Tian, A. Zhang, C. Qiu, W. Wang, Y. Yang, C. Qiu, et al., "The Upregulation of LAG-3 on T Cells Defines a Subpopulation with Functional Exhaustion and Correlates with Disease Progression in HIV-Infected Subjects" J. Immunol., vol. 194, pp. 3873-3882, 2015. [Crossref]
[39] M. Wang, Q. Du, J. Jin, Y. Wei, Y. Lu, Q. Li, "LAG3 and Its Emerging Role in Cancer Immunotherapy" Clin. Transl. Med., vol. 11, p. e365, 2021. [Crossref]
[40] M. Guo, F. Yuan, F. Qi, J. Sun, Q. Rao, Z. Zhao, et al., "Expression and Clinical Significance of LAG-3, FGL1, PD-L1 and CD8(+)T Cells in Hepatocellular Carcinoma Using Multiplex Quantitative Analysis" J. Transl. Med., vol. 18, p. 306, 2020. [Crossref]
[41] W.J. Lee, Y.J. Lee, M.E. Choi, K.A. Yun, C.H. Won, M.W. Lee, et al., "Expression of Lymphocyte-Activating Gene 3 and T-Cell Immunoreceptor with Immunoglobulin and ITIM Domains in Cutaneous Melanoma and Their Correlation with Programmed Cell Death 1 Expression in Tumor-Infiltrating Lymphocytes" J. Am. Acad. Dermatol., vol. 81, pp. 219-227, 2019. [Crossref]
[42] W.W. Deng, L. Mao, G.T. Yu, L.L. Bu, S.R. Ma, B. Liu, et al., "LAG-3 Confers Poor Prognosis and Its Blockade Reshapes Antitumor Response in Head and Neck Squamous Cell Carcinoma" Oncoimmunology, vol. 5, p. e1239005, 2016. [Crossref]
[43] K. Lv, R. Li, Y. Cao, Y. Gu, X. Liu, X. He, et al., "Lymphocyte-Activation Gene 3 Expression Associates with Poor Prognosis and Immunoevasive Contexture in Epstein-Barr Virus-Positive and MLH1-Defective Gastric Cancer Patients" Int. J. Cancer, vol. 148, pp. 759-768, 2021. [Crossref]
[44] G. Zhou, L. Noordam, D. Sprengers, M. Doukas, P.P.C. Boor, A.A. van Beek, et al., "Blockade of LAG3 Enhances Responses of Tumor-Infiltrating T Cells in Mismatch Repair-Proficient Liver Metastases of Colorectal Cancer" Oncoimmunology, vol. 7, p. e1448332, 2018. [Crossref]
[45] S. Burugu, D. Gao, S. Leung, S.K. Chia, T.O. Nielsen, "LAG-3+ Tumor Infiltrating Lymphocytes in Breast Cancer: Clinical Correlates and Association with PD-1/PD-L1+ Tumors" Ann. Oncol., vol. 28, pp. 2977-2984, 2017. [Crossref]
[46] S.M. Hald, M. Rakaee, I. Martinez, E. Richardsen, S. Al-Saad, E.E. Paulsen, et al., "LAG-3 in Non-Small-cell Lung Cancer: Expression in Primary Tumors and Metastatic Lymph Nodes Is Associated With Improved Survival" Clin. Lung Cancer, vol. 19, pp. 249-259.e242, 2018. [Crossref]
[47] Y. Zhang, Y.D. Liu, Y.L. Luo, B.L. Liu, Q.T. Huang, F. Wang, et al., "Prognostic Value of Lymphocyte Activation Gene-3 (LAG-3) Expression in Esophageal Squamous Cell Carcinoma" J. Cancer, vol. 9, pp. 4287-4293, 2018. [Crossref]
[48] Y. Sun, J. Luo, Y. Chen, J. Cui, Y. Lei, Y. Cui, et al., "Combined Evaluation of the Expression Status of CD155 and TIGIT Plays an Important Role in the Prognosis of LUAD (Lung Adenocarcinoma)" Int. Immunopharmacol., vol. 80, p. 106198, 2020. [Crossref]
[49] X. Liu, M. Li, X. Wang, Z. Dang, Y. Jiang, X. Wang, et al., "PD-1(+) TIGIT(+) CD8(+) T Cells Are Associated with Pathogenesis and Progression of Patients with Hepatitis B Virus-Related Hepatocellular Carcinoma" Cancer Immunol. Immunother. CII, vol. 68, pp. 2041-2054, 2019. [Crossref]
[50] R. Saleh, R.Z. Taha, S.M. Toor, V. Sasidharan Nair, K. Murshed, M. Khawar, et al., "Expression of Immune Checkpoints and T Cell Exhaustion Markers in Early and Advanced Stages of Colorectal Cancer" Cancer Immunol. Immunother. CII, vol. 69, pp. 1989-1999, 2020. [Crossref]
[51] C. Giampietri, F. Scatozza, E. Crecca, V. Vigiano Benedetti, P.G. Natali, A. Facchiano, "Analysis of Gene Expression Levels and Their Impact on Survival in 31 Cancer-Types Patients Identifies Novel Prognostic Markers and Suggests Unexplored Immunotherapy Treatment Options in a Wide Range of Malignancies" J. Transl. Med., vol. 20, p. 467, 2022. [Crossref] [PubMed]
[52] X. Zhou, X. Ding, H. Li, C. Yang, Z. Ma, G. Xu, et al., "Upregulation of TIGIT and PD-1 in Colorectal Cancer with Mismatch-Repair Deficiency" Immunol. Investig., vol. 50, pp. 338-355, 2021. [Crossref]
[53] R. Liang, X. Zhu, T. Lan, D. Ding, Z. Zheng, T. Chen, et al., "TIGIT Promotes CD8(+)T Cells Exhaustion and Predicts Poor Prognosis of Colorectal Cancer" Cancer Immunol. Immunother., vol. 70, pp. 2781-2793, 2021. [Crossref]
[54] M. Kitsou, G.D. Ayiomamitis, A. Zaravinos, "High Expression of Immune Checkpoints Is Associated with the TIL Load, Mutation Rate and Patient Survival in Colorectal Cancer" Int. J. Oncol., vol. 57, pp. 237-248, 2020. [Crossref]
[55] J.M. Chauvin, O. Pagliano, J. Fourcade, Z. Sun, H. Wang, C. Sander, et al., "TIGIT and PD-1 Impair Tumor Antigen-Specific CD8⁺ T Cells in Melanoma Patients" J. Clin. Investig., vol. 125, pp. 2046-2058, 2015. [Crossref]
[56] P. Wang, Y. Chen, Q. Long, Q. Li, J. Tian, T. Liu, et al., "Increased Coexpression of PD-L1 and TIM3/TIGIT Is Associated with Poor Overall Survival of Patients with Esophageal Squamous Cell Carcinoma" J. Immunother. Cancer, vol. 9, p. e002836, 2021. [Crossref]
[57] M. Nakano, M. Ito, R. Tanaka, K. Yamaguchi, H. Ariyama, K. Mitsugi, et al., "PD-1+ TIM-3+ T Cells in Malignant Ascites Predict Prognosis of Gastrointestinal Cancer" Cancer Sci., vol. 109, pp. 2986-2992, 2018. [Crossref]
[58] A. Alsalman, M.A. Al-Mterin, K. Murshed, F. Alloush, S.T. Al-Shouli, S.M. Toor, et al., "Circulating and Tumor-Infiltrating Immune Checkpoint-Expressing CD8(+) Treg/T Cell Subsets and Their Associations with Disease-Free Survival in Colorectal Cancer Patients" Cancers, vol. 14, 2022. [Crossref]
[59] J. Tan, S. Huang, J. Huang, Z. Yu, Y. Chen, Y. Lu, et al., "Increasing Tim-3+CD244+, Tim-3+CD57+, and Tim-3+PD-1+ T Cells in Patients with Acute Myeloid Leukemia" Asia-Pac. J. Clin. Oncol., vol. 16, pp. 137-141, 2020. [Crossref]
[60] J. Tan, Z. Yu, J. Huang, Y. Chen, S. Huang, D. Yao, et al., "Increased PD-1+Tim-3+ Exhausted T Cells in Bone Marrow May Influence the Clinical Outcome of Patients with AML" Biomark. Res., vol. 8, 2020. [Crossref]
[61] M. Klapholz, M.G. Drage, A. Srivastava, A.C. Anderson, "Presence of Tim3(+) and PD-1(+) CD8(+) T Cells Identifies Microsatellite Stable Colorectal Carcinomas with Immune Exhaustion and Distinct Clinicopathological Features" J. Pathol., vol. 257, pp. 186-197, 2022. [Crossref] [PubMed]
[62] N.E. Kushlinskii, O.V. Kovaleva, A.N. Gratchev, A.A. Alferov, Y.B. Kuzmin, N.Y. Sokolov, et al., "Assessing the Clinical Relevance of Soluble PD-1 and PD-L1: A Multi-Cohort Study Across Diverse Tumor Types and Prognostic Implications" Biomedicines, vol. 13, 2025. [Crossref] [PubMed]
[63] M. Dank, D. Mühl, M. Herold, L. Hornyák, A.M. Szasz, Z. Herold, "Does Elevated Pre-Treatment Plasma PD-L1 Level Indicate an Increased Tumor Burden and Worse Prognosis in Metastatic Colorectal Cancer?" J. Clin. Med., vol. 11, 2022. [Crossref] [PubMed]
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more