
APA Style
Manikant Tripathi, Neeraj Pathak, Gaurav Yadav, Sukriti Pathak, Pankaj Singh, Pradeep Kumar Singh, Diksha Garg. (2025). Effective Decolorization of Direct Blue-1 Dye by Bacillus cereus Isolate for Its Possible Application in Environmental Sustainability. Sustainable Processes Connect, 1 (Article ID: 0006). https://doi.org/10.69709/SusProc.2025.118221MLA Style
Manikant Tripathi, Neeraj Pathak, Gaurav Yadav, Sukriti Pathak, Pankaj Singh, Pradeep Kumar Singh, Diksha Garg. "Effective Decolorization of Direct Blue-1 Dye by Bacillus cereus Isolate for Its Possible Application in Environmental Sustainability". Sustainable Processes Connect, vol. 1, 2025, Article ID: 0006, https://doi.org/10.69709/SusProc.2025.118221.Chicago Style
Manikant Tripathi, Neeraj Pathak, Gaurav Yadav, Sukriti Pathak, Pankaj Singh, Pradeep Kumar Singh, Diksha Garg. 2025. "Effective Decolorization of Direct Blue-1 Dye by Bacillus cereus Isolate for Its Possible Application in Environmental Sustainability." Sustainable Processes Connect 1 (2025): 0006. https://doi.org/10.69709/SusProc.2025.118221.
 ACCESS
Research Article
ACCESS
Research Article
Volume 1, Article ID: 2025.0006

Manikant Tripathi
manikant.microbio@gmail.com

Neeraj Pathak
npathakniraj2000@gmail.com

Gaurav Yadav
gy6047969@gmail.com

Sukriti Pathak
sukritipathak24@gmail.com

Pankaj Singh
singhpankaj0984@rediffmail.com

Pradeep Kumar Singh
pkbt99@gmail.com

Diksha Garg
1 Biotechnology Program, Dr. Rammanohar Lohia Avadh University, Ayodhya 224001, Uttar Pradesh, India
2 Department of Biochemistry, Dr. Rammanohar Lohia Avadh University, Ayodhya 224001, Uttar Pradesh, India
3 Department of Microbiology, DAV University, Jalandhar 144012, Punjab, India
* Author to whom correspondence should be addressed
Received: 25 Feb 2025 Accepted: 27 Jul 2025 Available Online: 27 Jul 2025 Published: 05 Aug 2025
Azo dyes, commonly found in textile wastewater, are major environmental contaminants that pose threats to a healthy ecosystem. Rapid industrial development has led to a surge in dye-contaminated wastewater, raising global environmental concerns due to the complex and resistant structure of these dyes. Bacterial remediation of dyes from contaminated aquatic systems is an environmentally friendly approach. The aim of this study was to isolate and identify a bacterial strain with the potential to decolorize an azo dye, Direct Blue-1, from dye-laden wastewater, and optimization of process parameters to achieve maximum level of dye decolorization. A total of 9 bacterial cultures were isolated at 50 mg/l dye concentration from dye-contaminated wastewater. While a single bacterial isolate was capable of decolorizing Direct Blue-1 dye at 200 mg/l in minimal salt agar medium. Morphological, biochemical, and 16S-rRNA gene sequence analysis revealed that the isolated bacterium was Bacillus cereus strain MT-4. Under optimized culture conditions, a maximum of 97% decolorization was achieved with a 5% (v/v) inoculum. This study highlights the potential of Bacillus cereus MT-4 for the effective removal of Direct Blue-1, suggesting its application in sustainable wastewater treatment.

The study supports the eco-friendly and cost-effective potential of microbial remediation for dye-contaminated wastewater. Bacillus cereus MT-4 was capable of decolorizing Direct Blue-1 dye. Optimal conditions included pH 7.0, temperature 35 °C, inoculum size 5% v/v, dye concentration 50 mg/L, and 48 h incubation. Static culture conditions were more effective (94.7%) than shaking conditions (83%). Dye concentrations above 200 mg/L inhibited bacterial growth and decolorization efficiency.
Ecosystem pollution continues to be a significant global challenge, with dye-laden effluent emerging as one of the major concerns. The widespread utilization of dyes driven by accelerated industrial expansion and increasing demand in multiple sectors has severely aggravated the issue [1]. Approximately 5–10% of dyes are discharged into industrial effluent, with azo-dyes representing 60% of the total dyes utilized [2,3,4]. Currently, azo-dyes and anthraquinone dyes are the two primary classes utilized for fabric dyeing. Under typical conditions, azo dyes are resistant to degradation and not efficiently removed by the traditional wastewater treatment processes due to their complex structure and anthropogenic nature of azo- dyes. Some of the dyes and their breakdown products facilitate the spread of toxic groundwater and surface water contamination from industrial textile efflux, further affecting alternative water sources [5,6,7]. Various pollutants, including azo dyes, heavy metals, microplastics, and other emerging pollutants, are adversely affecting the ecosystem [8,9,10,11]. Azo-dyes present pose significant environmental and health concerns due to their tumorigenic, toxic, and mutagenic properties [12]. Direct Blue (DB) dye is used in textiles, and many researchers have studied the microbial-based remediation of DB dyes [13,14,15]. Azo-dyes and their degradation byproducts, including aromatic amines, are recognized as mutagenic and carcinogenic compounds [16,17]. Hernández-Zamora et al. [18] reported the toxic impacts of dye on microalgae, cladocerans, and embryos of zebrafish. Dye pollution also results in undesirable water coloration, hinders the penetration of light, and reduces the dissolved oxygen levels in the riverine habitat, threatening water-dwelling species. Therefore, proper treatment of dye-laden industrial effluent is crucial before its release into water bodies. Biological processes for the treatment of textile effluents are preferred over physicochemical methods due to their affordability, eco-friendly nature, and lower sludge production [19,20]. Microbe-based remediation is suitably employed for the mitigation of environmental pollutants [21,22]. Previous studies have reported remediation and decolorization of azo dyes from a contaminated environment [14,23]. As a sustainable solution for dye detoxification, microbe-mediated processes are being globally recognized. Bacterial remediation approach has been developed into a key strategy for treating industrial effluent due to its multiple advantages. Particularly, they can persist and develop substantial biomass even under the nutrient-limited conditions. Additionally, their potential in adapting to harsh environmental conditions through various facultative anaerobic, anaerobic, and aerobic pathways highlights their ecological effectiveness [24,25,26]. Researchers reported dye remediation by microbes like Di-Azo dye Direct Red 81 degraded by a bacterial mixed culture [27], and Direct blue-1 dye decolorization and degradation by the fungus Aspergillus terreus GS28 [14]. Optimization of different process factors needs to be studied for achieving the maximum level of dye decolorization. Moreover, the application of a single efficient bacterial isolate may be a cost-effective approach for on-site dye decolorization. In this study, a potential Direct Blue-1 decolorizing bacterium was isolated from dye wastewater and identified by morphological and molecular methods. The effect of potent strain on the degradation of Direct blue-1 was optimized at different temperatures for identifying the maximum level of dye decolorization for its possible role in environmental bioremediation.
2.1. Sampling Dye-contaminated wastewater samples were collected in sterile bottles near the discharge points of power looms in Tanda, Ambedkar Nagar, Uttar Pradesh, India, and stored at 4 °C for further experimentation. 2.2. Isolation and Screening of Potential Direct Blue 1 Dye Decolorizing Bacteria The pour plate technique was employed to isolate bacteria (at 35 °C for 48 h), which were capable of decolorizing Direct blue-1 on solid minimum salt medium (MSM) containing 50 mg dye concentration. For the screening of the most potent Direct blue-1 dye decolorizing bacterium and its further application in decolorization studies, MSM agar plates were added with altered with dye concentration (50–300 mg/L), incubated, and observed for further study. 2.3. Identification of Potential Direct Blue 1 Dye Decolorizing Bacterium Morphological and biochemical characteristics were used for preliminary identification [28]. Additionally, molecular characterization was used to identify bacteria at the species level by using the 16S rRNA gene sequencing application. The 16S rRNA gene sequence analysis was carried out at CytoGene Research & Development, Lucknow (India). 2.4. Culture Conditions for Dye Decolorization The dye decolorization trials were performed in the broth media, and inoculated with selected potential bacterial isolates, and incubated at 35 °C for 72 h. All experiments were conducted in triplicate. Periodically, the media samples were taken out and the extent of dye discoloration was examined at every 24 h, and analysed spectrophotometrically (at 593 nm). 2.5. Factors Various process factors influencing bacterial dye decolorization efficiency were studied to determine optimal conditions. 2.5.1. Effect of Static and Shaking of Culture Medium In order to determine the impact of shaking and static culture conditions, the extent of dye discoloration was measured at 48 h incubation. 2.5.2. Effect of Dye Concentration, Incubation Time, pH, Temperature, and Inoculum Dose The impact of different parameters like dye level (50–200 mg/L), pH (5.0–9.0), temperature (25–40 °C), incubation time (24–72 h), and inoculum dose (1–4% v/v) on discoloration of Direct blue 1 dye was investigated. Samples were treated under the previously described culture conditions. 2.6. Measurement for the Extent of Dye Discoloration To determine Direct blue-1 dye discoloration, the samples drawn every 24 h were centrifuged (at 10,000 rpm for 10 min). Further, the supernatant was analysed to determine the percentage of decolorization by using a UV-vis spectrophotometer (Labtronics) [14,29]. The extent of dye discoloration was calculated using the following formula: 2.6.1. Statistical Analyses The bacterial decolorization experiments were executed in triplicate and the statistical analysis (standard deviation) was done by using the Microsoft Excel program.
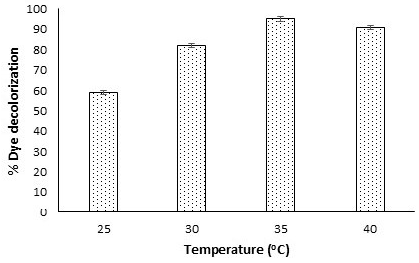
The improper release of coloured wastewater to the aquatic environment is one of the significant concerns. The presence of azo dyes in the aquatic ecosystems has adverse effects on humans, animals, and plants. Microbe-based decolorization is one of the viable options that helps in environmental sustainability by removing dye pollution. To facilitate bacterial biodecolorization, optimization of process parameters was executed. 3.1. Isolation of Potential Bacteria for Discoloration of Direct Blue-1 Dye To isolate dye-decolorizing bacteria, the sample was inoculated on MSM agar medium. In this study, the dye was decolorized in the medium after 48 h of incubation (Figure 1). Further, the bacterial colonies were isolated and purified on the same medium. A total of nine bacterial isolates were found to be capable of decolorizing Direct blue 1 dye at 50 mg/L concentration within 48 h of incubation. While, single bacterial isolate was capable of growing on MSA medium amended with 200 mg/L dye concentration (Table 1). There was no growth of any bacterial isolates above 200 mg/L dye concentration. Higher concentration of dye (above 200 mg/L of Direct Blue 1) were inhibitory for bacterial growth. Several researchers studied the effect of dye level on the extent of microbe-based decolorization [23,30,31,32]. In a research investigation, dye decolorization was assessed using mixed consortia of Bacillus sp., at different dye concentrations from 200 to 1000 mg/l, and reported decreased extent of decolorization with increasing dye concentration [33]. The increasing concentration of dye might be toxic to bacterial isolates, resulting in reduced dye decolorization. Screening of the most potential Direct blue-1 decolorizing bacterium. 3.2. Morphological, Biochemical 16S rRNA Gene Sequence Analyses for Identification of Potential Bacterial Isolate In this study, a potential Direct blue-1 decolorizing bacterial, NSW-1, was isolated. Morphologically, the bacterial isolate was rod-shaped, and biochemical characterization showed the isolate was Gram-positive, catalase-positive, and capable of utilizing carbohydrates like glucose, sucrose, etc. Further, the isolate NSW-1 was identified as Bacillus cereus MT-4 using 16S rRNA gene sequence analysis. Figures S1 and S2 (supplementary) show the gel images of isolated genomic DNA and PCR-amplified products. The experiments for DNA extraction, agarose gel electrophoresis, and PCR were conducted at Cytogene Research Lab, Lucknow. The accession number LC830210 was assigned after depositing the 16S rRNA gene sequence (1470 bp) to the DNA Data Bank of Japan database (https://www.ncbi.nlm.nih.gov/nuccore/LC830210.1/, access date 9 July 2024). The phylogenetic tree was constructed to show the relatedness with other related genera (Figure 2). 3.3. Optimization of Factors Affecting Dye Decolorization Different biodecolorization conditions like dye concentration, temperature, incubation time, pH, culture medium shaking, and inoculum size were optimized. Biodecolorization of Direct blue-1 was better under static conditions (94.7%) compared to shaking conditions (83%) at 48 h incubation. This might be due to better enzymatic activity under no-shaking culture conditions. While the percent decolorization was lower at 24 h, an insignificant change in percent decolorization was observed after 48 h of incubation. Dye decolorization efficiency was attempted with varying dye concentrations from 50 to 200 mg/l. It was observed that maximum decolorization was achieved at 50 mg/L; further increases in dye concentration caused a decrease in the percentage of decolorization. A minimum of 17% decolorization was observed at 200 mg/l concentration. In this study, the extent of dye decolorization was calculated using optical density (OD) measurement. Other researchers also used OD for measuring decolorization of dyes [6,34]. In a research investigation, Afrin et al. [34] reported degradation of textile dyes using a bacterial consortium by measuring OD at optimized conditions, pH 7.0, 37 °C, at 100 mg/L dye concentration. Further, Direct blue-1 (50 mg/L) amended medium for bacterial decolorization was exposed to a wide range of temperatures of 25–40 °C. It was observed that temperature alteration can significantly impact the bacterial decolorization of azo dyes. Approximately 95% of decolorization of Direct Blue-1 dye was observed by using 4% v/v inoculum dose at pH 7.0 and 35 °C. There was a decrease in decolorization at various other tested temperatures (Figure 3). Further decolorization studies were attempted at 48 h incubation and 35 °C. Previous studies by multiple researchers, such as El Awady et al. [35], observed the optimal decolorization of different functional azo dyes by Streptomyces albidoflavus 3MGH around the temperature of 35 °C. Similarly, Barathi et al. [7], reported a similar trend of maximum dye decolorization at 35 °C of Reactive Blue 160 textile dye by Bacillus subtilis. In another investigation, Srivastava et al. [36] found efficient decolorization at 40 °C. These findings suggest that at 35–40 °C, bacteria exhibit significant decolorization activity, likely due to enhanced enzymatic performance. Several researchers studied on decolorization of dyes for environmental sustainability and pollutant remediation [6,7,13]. The extent of dye degradation was significantly affected by the amount of bacterial inoculum. The range of inoculum dose taken into account for the investigation of Direct Blue-1 dye decolorization was 1.0–6.0% v/v. At 5% of inoculum size, the decolorization was found to be a maximum of 97%, while a decreased percent decolorization was reported with any deviation from the optimal value of inoculum dose. In the present study, the results showed that with an increase in the size of the inoculum, the decolorization of dye rises, which may be due to higher enzyme production. In a study, Srivastava et al. [36] studied biodecolorization of Reactive Black 5 dye by B. albus DD1 isolated from textile water effluent, and they found the highest decolorization (74.7 ± 2.1%) with a higher inoculum size (25% v/v), while lesser decolorization (54.9 ± 1.2%) with 5% inoculum size. However, in this study, no significant change in decolorization was observed by increasing inoculum size from 5% to 6%, which might be due to depletion of nutrients because of increased biomass, which may result in lesser enzymatic function [36,37]. pH is one of the crucial factors for any bioprocess and significantly influences microbial functions. The pH range for the Direct Blue-1 dye decolorization was from 5 to 9. It was observed that at pH 7, maximum decolorization (97%) of the dye was obtained. Further deviation in pH from the optimal value resulted in decreased biodecolorization. Similarly, the optimum pH for maximum decolorization by Bacillus alba was reported to be 7.0 by Srivastava et al [36] in their investigation. In a study, Neetha et al. [38] optimized decolorization of Direct Blue-14 dye by B. fermus isolate. In another investigation, El Bouraie & El Din [39] also observed the maximum result for dye decolorization at pH 7. The findings suggest that the bacterium Bacillus sp. prefers neutral pH for its maximum decolorization capability. Many researchers also investigated the decolorization and detoxification of Direct blue dye by bacteria [40,41,42]. Wu et al. [43] studied the enzymatic discoloration of methylene blue dye by Bacillus thuringiensis. These studies indicate bacterial-based decolorization may be an effective way for the remediation of dyes from a contaminated aquatic environment.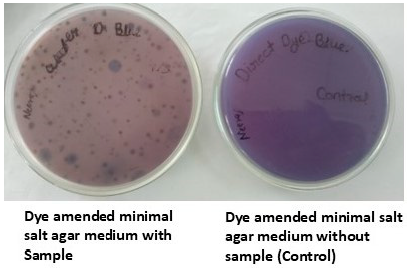
Sr. No.
Dye Concentration (mg/L)
Number of Bacterial Isolates
1
50
9
2
100
5
3
150
2
4
200
1
5
250
-
6
300
-

Addressing dye contamination is a critical requirement today. Different bioremediation strategies can be highly effective in the successful breakdown of synthetic dyes. In this study, the maximum decolorization of 97% was obtained by using a single culture of the bacterium Bacillus cereus MT-4 strain under optimized cultural conditions, pH, temperature, inoculum size, and dye concentration were 7.0, 35 °C, 4.0% and 50 mg/L dye, respectively. Further research investigations are required for the possible large-scale decolorization of Direct Blue-1 dye by an isolated bacterium.
| DB | Direct Blue |
| MSA | Minimal Salt Agar |
| MSM | Minimum Salt Medium |
| OD | Optical Density |
| PCR | Polymerase Chain Reaction |
| 16S rRNA | 16S ribosomal Ribonucleic Acid |
M.T.: Conceptualization, writing—review and editing, supervision; N.P.: Investigation, writing—original draft preparation; G.Y.: Investigation, writing—original draft preparation; S.P.: Writing—original draft preparation; P.S.: Writing—review and editing; P.K.S.: Writing—review and editing; D.G.: Writing—review and editing. All authors have read and agreed to the published version of the manuscript.
The data supporting the findings of this study are available within the article.
No consent for publication is required, as the manuscript does not involve any individual personal data, images, videos, or other materials that would necessitate consent.
All authors declare no conflicts of interest regarding this manuscript.
The study did not receive any external funding and was conducted using only institutional resources.
All authors are thankful to their parent universities for providing a research environment.
Download supplementary material Figure S1 and S2
[1] Abilaji, S.; Sathishkumar, K.; Narenkumar, J.; Alsalhi, M.S.; Devanesan, S.; Parthipan, P.; Muthuraj, B.; Rajasekar, A. Sequential Photo Electro Oxidation and Biodegradation of Textile Effluent: Elucidation of Degradation Mechanism and Bacterial Diversity. Chemosphere 2023, 331, 138816. [CrossRef] [PubMed]
[2] Yagub, M.T.; Sen, T.K.; Ang, H.M. Equilibrium, Kinetics, and Thermodynamics of Methylene Blue Adsorption by Pine Tree Leaves. Water Air Soil. Pollut. 2012, 223, 5267–5282. [CrossRef]
[3] Yagub, M.T.; Sen, T.K.; Afroze, S.; Ang, H.M. Dye and Its Removal from Aqueous Solution by Adsorption: A Review. Adv. Colloid Interface Sci. 2014, 209, 172–184. [CrossRef]
[4] Shah, M.P. Microbe-mediated degradation of synthetic dyes in wastewater. Microbial Degradation of Synthetic Dyes in Wastewaters ; Springer International Publishing: Cham, Switzerland, 2014; 205–241. . [CrossRef]
[5] Franciscon, E.; Piubeli, F.; Fantinatti-Garboggini, F.; de Menezes, C.R.; Silva, I.S.; Cavaco-Paulo, A.; Grossman, M.J.; Durrant, L.R. Polymerization Study of the Aromatic Amines Generated by the Biodegradation of Azo Dyes Using the Laccase Enzyme. Enzym. Microb. Technol. 2010, 46, 360–365. [CrossRef]
[6] Garg, S.K.; Tripathi, M.; Singh, S.K.; Tiwari, J.K. Biodecolorization of Textile Dye Effluent by Pseudomonas putida SKG-1 (MTCC 10510) Under the Conditions Optimized for Monoazo Dye Orange II Color Removal in Simulated Minimal Salt Medium. Int. Biodeterior. Biodegrad. 2012, 74, 24–35. [CrossRef]
[7] Barathi, S.; Aruljothi, K.N.; Karthik, C.; Padikasan, I.A. Optimization for Enhanced Eco-Friendly Decolorization and Detoxification of Reactive Blue160 Textile Dye by Bacillus subtilis. Biotechnol. Rep. 2020, 28. [CrossRef]
[8] Bala, S.; Garg, D.; Thirumalesh, B.V.; Sharma, M.; Sridhar, K.; Inbaraj, B.S.; Tripathi, M. Recent Strategies for Bioremediation of Emerging Pollutants: A Review for a Green and Sustainable Environment. Toxics 2022, 10. [CrossRef]
[9] Tripathi, M.; Shukla, S.; Singh, R.; Singh, S.; Singh, P.; Singh, P.K.; Shukla, A.K.; Maurya, S.; Pathak, S.; Chaudhary, V.K.; et al. Physicochemical Investigations of Textile Wastewater and Process Parameter Optimization for Biodecolorization of Congo Red Dye by Pseudomonas aeruginosa MT-2 Strain. J. Pure Appl. Microbiol. 2024, 18, 2558–2569. [CrossRef]
[10] Tripathi, M.; Singh, R.; Lal, B.; Haque, S.; Ahmad, I.; Yadav, A.K. Marine Microbial Bioremediation of Heavy Metal Contaminants in Waste Water for Health and Environmental Sustainability: A Review. Indian J. Microbiol. 2024, 65, 573–582. [CrossRef] [PubMed]
[11] Tripathi, M.; Singh, P.; Pathak, S.; Manimekalai, R.; Garg, D.; Dashora, K. Strategies for the Remediation of Micro- and Nanoplastics from Contaminated Food and Water: Advancements and Challenges. J. Xenobiotics 2025, 15. [CrossRef]
[12] Elisangela, F.; Andrea, Z.; Fabio, D.G.; de Menezes Cristiano, R.; Regina, D.L.; Artur, C.P. Biodegradation of Textile Azo Dyes by a Facultative Staphylococcus arlettae Strain VN-11 Using a Sequential Microaerophilic/Aerobic Process. Int. Biodeterior. Biodegrad. 2009, 63, 280–288. [CrossRef]
[13] Arun Prasad, A.S.; Satyanarayana, V.S.; Bhaskara Rao, K.V. Biotransformation of Direct Blue 1 by a Moderately Halophilic Bacterium Marinobacter sp. Strain HBRA and Toxicity Assessment of Degraded Metabolites. J. Hazard. Mater. 2013, 262, 674–684. [CrossRef] [PubMed]
[14] Singh, G.; Dwivedi, S.K. Decolorization and Degradation of Direct Blue-1 (Azo dye) by Newly Isolated Fungus Aspergillus terreus GS28, from Sludge of Carpet Industry. Environ. Technol. Innov. 2020, 18, 100751. [CrossRef]
[15] Ekanayake, E.M.M.S.; Manage, P.M. Green Approach for Decolorization and Detoxification of Textile Dye- CI Direct Blue 201 Using Native Bacterial Strains. Environ. Nat. Resour. J. 2020, 18, 1–8. [CrossRef]
[16] Saratale, R.G.; Saratale, G.D.; Kalyani, D.C.; Chang, J.S.; Govindwar, S.P. Enhanced Decolorization and Biodegradation of Textile Azo Dye Scarlet R By using Developed Microbial Consortium-GR. Bioresour. Technol. 2009, 100, 2493–2500. [CrossRef]
[17] Ebency, C.I.L.; Rajan, S.; Murugesan, A.G.; Rajesh, R.; Elayarajah, B. Biodegradation of Textile Azo Dyes and Its Bioremediation Potential Using Seed Germination Efficiency. Int. J. Curr. Microbiol. App. Sci. 2013, 2, 496–505.Available online: https://www.ijcmas.com/vol-2-10/C.Initha%20Lebanon%20Ebency,%20et%20al.pdf.
[18] Hernández-Zamora, M.; Martínez-Jerónimo, F. Exposure to the Azo Dye Direct Blue 15 Produces Toxic Effects on Microalgae, Cladocerans, and Zebrafish Embryos. Ecotoxicology 2019, 28, 890–902. [CrossRef] [PubMed]
[19] Gurulakshmi, M.; Sudarmani, D.N.P.; Venba, R. Biodegradation of Leather Acid Dye by Bacillus subtilis. Adv. Biotech. 2008, 7, 12–19.Available online: https://www.scribd.com/document/49992433/59-Biodegradation-of-Leather.
[20] Hassan, M.M.; Alam, M.Z.; Anwar, M.N. Biodegradation of Textile Azo Dyes by Bacteria Isolated from Dyeing Industry Effluent. Int. Res. J. Biol. Sci. 2013, 2, 27–31.Available online: https://www.isca.me/IJBS/Archive/v2/i8/6.ISCA-IRJBS-2013-098.pdf.
[21] Gaur, R.; Bala, S.; Tripathi, M.; Kumar, R. Microbial Biofilms: Revolutionizing Fermentation and Bioremediation of Environmental Pollutants. In Challenges and Sustainable Solutions in Bioremediation , Dubey, K.K.; Yadav, A.; Shah, M.P., Eds.; CRC Press: Boca Raton, FL, USA, 2025; 17–33. Available online: https://www.taylorfrancis.com/chapters/edit/10.1201/9781003407317-2/microbial-biofilms-rajeeva-gaur-saroj-bala-manikant-tripathi-ravi-kumar..
[22] Gupta, A.; Khan, F.; Pandey, P.; Tripathi, M.; Pathak, N. A Comprehensive Review on the Role of Biosurfactants in Remediation of Heavy Metals from Contaminated Environment. Bioremed. J. 2024, , 1–27. [CrossRef]
[23] Tripathi, M.; Pathak, N.; Chaudhary, V.K.; Singh, P.; Singh, P.K.; Thirumalesh, B.V.; Bala, S.; Maurya, A.K.; Patel, N.; Yadav, B.K. Microbial Decolorization of Crystal Violet Dye by a Native Multi-Metal Tolerant Aeromonas caviae MT-1 Isolate from Dye-Contaminated Soil: Optimization and Phytotoxicity Study. Toxicol. Int. 2023, 30, 83–93. [CrossRef]
[24] Olukanni, O.D.; Osuntoki, A.A.; Kalyani, D.C.; Gbenle, G.O.; Govindwar, S.P. Decolorization and biodegradation of Reactive Blue 13 by Proteus mirabilis LAG. J. Hazard. Mater. 2010, 184, 290–298. [CrossRef] [PubMed]
[25] Sharma, R.; Kumar, N.; Sharma, P.; Yadav, A.; Aggarwal, N.K. Biological Decolorisation of the Anionic Dye Acid Blue 9 by Bacterial Consortium: A Sustainable and Ecofriendly Approach for the Treatment of Textile Wastewater. Sustain. Chem. Environ. 2024, 8, 100178.Available online: https://www.scribd.com/document/725333629/Bergey-s-Manual-of-Determinative-Bacteriology-Bergey-D-H-David. [CrossRef]
[26] Ravi, A.; Krishnan, R.; Ravuri, M.; Santhosh, S.; AlSalhi, M.S.; Devanesan, S.; Selvarani, A.; Rajasekar, A.; Rajamohan, R.; Narenkumar, J. Sustainable Approach for the Degradation of Contrast Dye Evans Blue by Enterobacter Cloacae Strain SD4-1. J. Taiwan. Inst. Chem. Eng. 2025, 166, 105323. [CrossRef]
[27] Kamal, I.M.; Abdeltawab, N.F.; Ragab, Y.M.; Farag, M.A.; Ramadan, M.A. Biodegradation, Decolorization, and Detoxification of Di-Azo Dye Direct Red 81 by Halotolerant, Alkali-Thermo-Tolerant Bacterial Mixed Cultures. Microorganisms 2022, 10. [CrossRef]
[28] Holt, J.G.; Krieg, N.R.; Sneath, P.H.A.; Staley, J.T.; Williams, S.T. . Bergey’s Manual of Determinative Bacteriology ; Williams & Wilkins: Baltimore, MD, USA, 1994; . .
[29] He, X.L.; Song, C.; Li, Y.; Wang, N.; Xu, L.; Han, X.; Wei, D. Efficient Degradation of Azo Dyes by a New Isolated Fungus Trichoderma tomentosum Under Non-Sterile Conditions. Ecotoxicol. Environ. Saf. 2018, 150, 232–239. [CrossRef]
[30] Seyedi, Z.S.; Zahraei, Z.; Jookar Kashi, F. Decolorization of Reactive Black 5 and Reactive Red 152 Azo Dyes by New Haloalkaliphilic Bacteria Isolated from the Textile Wastewater. Curr. Microbiol. 2020, 77, 2084–2092. [CrossRef]
[31] Sharma, R.; Sharma, P.; Kumar, N.; Aggarwal, N.K. Enhanced Decolorization of Basic Violet 14 Dye Wastewater and Its Phytotoxicity Assessment: A Green Approach. Environ. Qual. Manag. 2025, 34, e70050. [CrossRef]
[32] Tripathi, M.; Singh, S.; Pathak, S.; Kasaudhan, J.; Mishra, A.; Bala, S.; Garg, D.; Singh, R.; Singh, P.; Singh, P.K.; et al. Recent Strategies for the Remediation of Textile Dyes from Wastewater: A Systematic Review. Toxics 2023, 11. [CrossRef]
[33] Bhoosreddy, G.L. Decolorization and Biodegradation of Direct Blue 2B by Mix Consortia of Bacillus. IOSR J. Pharm. Biol. Sci. 2014, 9, 34–40. [CrossRef]
[34] Afrin, S.; Shuvo, H.R.; Sultana, B.; Islam, F.; Rus’d, A.A.; Begum, S.; Hossain, M.N. The Degradation of Textile Industry Dyes Using the Effective Bacterial Consortium. Heliyon 2021, 7, e08102. [CrossRef]
[35] El Awady, M.E.; El-Shall, F.N.; Mohamed, G.E.; Abd-Elaziz, A.M.; Abdel-Monem, M.O.; Hassan, M.G. Exploring the Decolorization Efficiency and Biodegradation Mechanisms of Different Functional Textile Azo Dyes by Streptomyces albidoflavus 3MGH. BMC Microbiol. 2024, 24. [CrossRef]
[36] Srivastava, A.; Dangi, L.K.; Kumar, S.; Rani, R. Microbial Decolorization of Reactive Black 5 dye by Bacillus Albus DD1 Isolated from Textile Water Effluent: Kinetic, Thermodynamics & Decolorization Mechanism. Heliyon 2022, 8, e08834. [CrossRef]
[37] Bonugli-Santos, R.C.; Vieira, G.A.; Collins, C.; Fernandes, T.C.C.; Marin-Morales, M.A.; Murray, P.; Sette, L.D. Enhanced Textile Dye Decolorization by Marine-Derived Basidiomycete Peniophora sp. CBMAI 1063 Using Integrated Statistical Design. Environ. Sci. Pollut. Res. 2016, 23, 8659–8668. [CrossRef] [PubMed]
[38] Neetha, J.N.; Sandesh, K.; Girish Kumar, K.; Chidananda, B.; Ujwal, P. Optimization of Direct Blue-14 Dye Degradation by Bacillus fermus (Kx898362) an Alkaliphilic Plant Endophyte and Assessment of Degraded Metabolite Toxicity. J. Hazard. Mater. 2019, 364, 742–751. [CrossRef]
[39] El Bouraie, M.; El Din, W.S. Biodegradation of Reactive Black 5 by Aeromonas hydrophila strain Isolated from Dye-Contaminated Textile Wastewater. Sustain. Environ. Res. 2016, 26, 209–216. [CrossRef]
[40] Cao, J.; Sanganyado, E.; Liu, W.; Zhang, W.; Liu, Y. Decolorization and Detoxification of Direct Blue 2B by Indigenous Bacterial Consortium. J. Environ. Manag. 2019, 242, 229–237. [CrossRef]
[41] Dang, S.; Fan, W.; Meng, F.; Li, X.; Hao, J.; Wang, C. Decolorization and Detoxification of Direct Blue 5B by a Marinobacter-Dominated Halo-Thermoalkalophilic Consortium. Chemosphere 2024, 363, 142957. [CrossRef]
[42] Lalnunhlimi, S.; Krishnaswamy, V. Decolorization of azo dyes (Direct Blue 151 and Direct Red 31) by Moderately Alkaliphilic Bacterial Consortium. Braz. J. Microbiol. 2016, 47, 39–46. [CrossRef]
[43] Wu, K.; Shi, M.; Pan, X.; Zhang, J.; Zhang, X.; Shen, T.; Tian, Y. Decolourization and Biodegradation of Methylene Blue Dye by a Ligninolytic Enzyme-Producing Bacillus thuringiensis: Degradation Products and Pathway. Enzym. Microb. Technol. 2022, 156, 109999. [CrossRef] [PubMed]
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more