
APA Style
Tammanna Ravee Sahrawat. (2025). Identification of Hub Genes and Molecular Mechanisms of Association of PCOS, Breast, and Ovarian Cancers Using an Integrated Bioinformatics Analysis. GenoMed Connect, 2 (Article ID: 0013). https://doi.org/10.69709/GenomC.2025.116635MLA Style
Tammanna Ravee Sahrawat. "Identification of Hub Genes and Molecular Mechanisms of Association of PCOS, Breast, and Ovarian Cancers Using an Integrated Bioinformatics Analysis". GenoMed Connect, vol. 2, 2025, Article ID: 0013, https://doi.org/10.69709/GenomC.2025.116635.Chicago Style
Tammanna Ravee Sahrawat. 2025. "Identification of Hub Genes and Molecular Mechanisms of Association of PCOS, Breast, and Ovarian Cancers Using an Integrated Bioinformatics Analysis." GenoMed Connect 2 (2025): 0013. https://doi.org/10.69709/GenomC.2025.116635.
 ACCESS
Research Article
ACCESS
Research Article
Volume 2, Article ID: 2025.0013

Tammanna Ravee Sahrawat
tammanna@pu.ac.in
1 Centre for Systems Biology and Bioinformatics, UIEAST, Panjab University, Chandigarh 160014, India
Received: 15 Dec 2024 Accepted: 28 Feb 2025 Available Online: 10 Mar 2025 Published: 21 Apr 2025
Background: Oxidative stress plays a crucial role in various aspects of cancer and other diseases. While reactive oxygen species (ROS) serve as key signal molecules in physiological processes for the normal functioning of the female reproductive system, they have also been implicated in pathological processes such as polycystic ovary syndrome (PCOS). Some studies have reported a significantly higher risk of endometrial cancer in women with PCOS. However, the association of PCOS and common female cancers, such as breast and ovarian cancer, has not been thoroughly studied. Objective: The present study was undertaken to identify the hub genes and molecular pathways that are common amongst polycystic ovary syndrome, breast cancer, and ovarian cancers using bioinformatics based on the interactomes of these diseases. Methodology: Common differentially expressed genes (DEGs) of PCOS, breast, and ovarian cancer were retrieved from GEO to analyze datasets using R-software. An interactome of the common DEGs and their interacting partners was built in the STRING database, followed by analysis using different Cytoscape plugins to identify and validate the hub genes and their functional enrichment. Results: The identified hub proteins, namely CYBA, CYBB, DUOX1, NCF1, NCF2, NCF4, NOX1, NOX3, NOXA1, and NOXO1, are components or regulators of the NADPH oxidase, which catalyzes the production of ROS that could promote carcinogenesis and metastasis in patients suffering from PCOS. Conclusions: NOX-derived ROS are essential for normal cellular functions and host defense against pathogens. However, excessive ROS production can lead to oxidative stress, contributing to various diseases, including PCOS and cancers. Therefore, regulating NADPH oxidase activity could potentially serve as a therapeutic approach for PCOS management and prevent the initiation and progression of cancers in females suffering from PCOS.
Oxidative stress [OS] occurs when the level of reactive oxygen species (ROS) exceeds the antioxidant capacity of the cell, resulting in damage to the cell and its components. ROS have a dual role in cellular function. At low concentrations, they act as signaling molecules involved in survival pathways essential for physiological functions and redox biology [1,2]. On the other hand, ROS are detrimental at high concentrations, leading to structural damage of nucleic acids, proteins, and lipids, thereby contributing to the pathogenesis of various diseases such as cardiovascular, neurodegenerative, and cancer [3]. Oxidative stress can affect various aspects of cancer, from initiation to treatment resistance, and is associated with both exogenous and endogenous factors [4]. ROS have been found to have a dual role, as it can induce senescence and apoptosis in cancer cells, thereby exhibiting antitumorigenic effects [5,6]. On the other hand, it can promote tumor growth by stimulating cell proliferation and angiogenesis, increasing DNA mutation rates, inducing genome instability, and modulating gene expression [7]. Prolonged exposure to high amounts of ROS can lead to the accumulation of DNA mutations due to the inhibition of DNA repair mechanisms, resulting in cancers [8]. ROS play a crucial role in the development and progression of breast and ovarian cancer. At moderate levels, they promote cancer cell survival by activating growth factors and signaling pathways [9,10]. NADPH oxidase enzymes play a significant role in breast cancer progression, with NOX1, NOX2, and NOX4 showing higher expression in breast cancer and DUOX1 levels being lower across all subtypes [11]. NOX1 is also highly expressed in ovarian tumors and is regulated by mitochondrial function, indicating a cross-talk between mitochondria and NOX signaling [12]. NOX4 has been reported to be over-expressed in ovarian tumors, contributing to tumorigenesis by promoting cellular senescence and apoptosis resistance [13,14]. Elevated levels of malondialdehyde (MDA) and reactive carbonyl proteins, both indicators of oxidative stress, have been observed in ovarian cancer patients, while levels of glutathione peroxidase 3 (GPX3) were found to be decreased [15]. As a secondary messenger, ROS can assist in tumor progression through the proliferation and survival of tumor cells by facilitating pro-tumorigenic signaling in the tumor microenvironment, thereby playing a crucial role in cancer invasion and metastasis [16]. A crosstalk between malignant and non-malignant cells results in this highly complex, multistep process leading to cancer metastasis [17,18]. Reactive oxygen species play a dual role in female reproduction, with both physiological and pathological effects, and the female reproductive tract contains multiple sources of ROS, such as Graafian follicles, follicular fluid, and the endometrium. At controlled levels, ROS are essential for various reproductive processes, including follicular development, ovulation, corpus luteum function, and embryo implantation. However, when ROS production exceeds antioxidant defenses, oxidative stress occurs, potentially damaging cells and tissues in the genital tract. This imbalance can negatively impact ovarian function and contribute to female infertility [19,20,21]. Recently, OS has been reported to play a significant role in the pathophysiology of polycystic ovary syndrome (PCOS) [22]. PCOS is a common endocrine disorder affecting up to 15–20% of reproductive-aged women, characterized by hyperandrogenism, oligo-anovulation, and polycystic ovaries [23]. Women with PCOS exhibit increased NADPH oxidase activity, a key enzyme in ROS production, which is associated with insulin resistance, abdominal adiposity, and hyperandrogenism [24]. Elevated ROS levels negatively impact ovarian follicles, oocyte quality, and fertility, resulting from mitochondrial dysfunction and impaired oxidative phosphorylation. This contributes to metabolic and hormonal dysregulation, as hyperandrogenism and insulin resistance create a feedback loop that further exacerbates oxidative stress in PCOS [25]. Multiple studies in women with PCOS have consistently found a significantly higher risk of endometrial cancer, which has been attributed to chronic anovulation leading to estrogen-driven endometrial hyperplasia [26,27]. OS plays a crucial role in breast cancer (BC) and ovarian cancer (OC) pathogenesis, contributing to its development, progression, and metastasis [6,10,28]. However, the relationship between PCOS and ovarian or breast cancer remains unclear. While some studies have found no significant association, others have suggested a potential link, highlighting the need for further investigation [27,29,30,31,32]. In view of the inconsistency in findings, the present study was undertaken to identify the molecular factors and pathways common amongst polycystic ovary syndrome and breast and ovarian cancer so as to discern their shared molecular signatures using an interactome-based bioinformatics approach.
2.1. Research Methodology The schematic workflow summarizing the bioinformatics pipeline is given in Figure 1. 2.2. Data Retrieval Datasets for the control and disease conditions of polycystic ovary syndrome (PCOS), breast (BC) and ovarian cancer (OC) having identifiers GSE84958, GSE42568, and GSE18520 [33,34,35], respectively, were retrieved from the NCBI Gene Expression Omnibus (GEO) database using the GEO query package of the R programming language [36]. 2.3. Pre-Processing of Microarray Data and DEG Identification The three GEO datasets were preprocessed through multiple steps, including data consolidation and normalization using log2 transformation, with further analysis performed using the gprofiler2 package in R Studio [37]. Volcano plots for all three datasets were constructed and visualized using the ggplot2 package [38]. To identify differential gene expression between disease and control states, up-regulated and down-regulated genes were obtained with p-value < 0.05 and logFC > 1, and p-value < 0.05 and logFC < −1, respectively, utilizing the limma package of R [39]. The overlaps and intersections among the three datasets of differentially expressed genes (DEGs) obtained for PCOS, BC, and OC were identified using Venny, an online tool that generates Venn diagrams [40]. 2.4. PPI Network Construction, Analysis, and Gene Enrichment Interactome to visualize the protein-protein interactions (PPI) was constructed using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) [41] and analyzed using Cytoscape [42]. Topological analysis and identification of functional modules of the networks was performed using the Cytoscape plug-in Cytocluster [43], MCODE [44], and NetworkAnalyst [45], along with CytoHubba to identify essential nodes/hubs using graph theory algorithms [46]. ClueGO, a Cytoscape plug-in, was used for pathway enrichment to identify the functional annotations [47].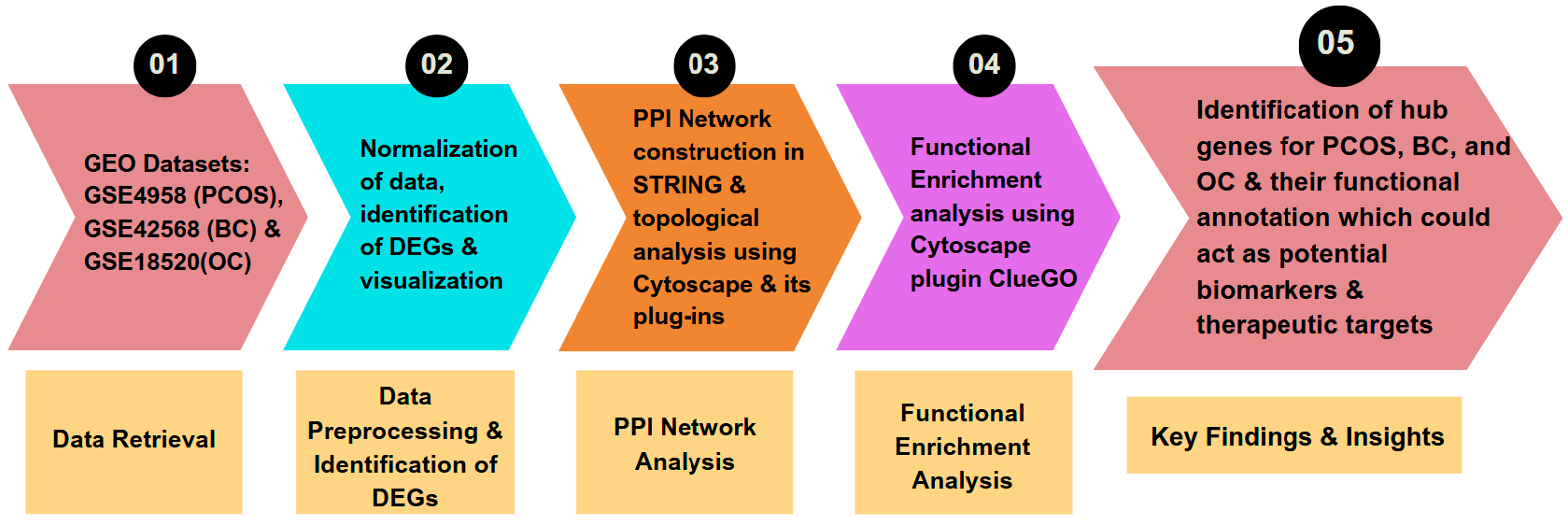
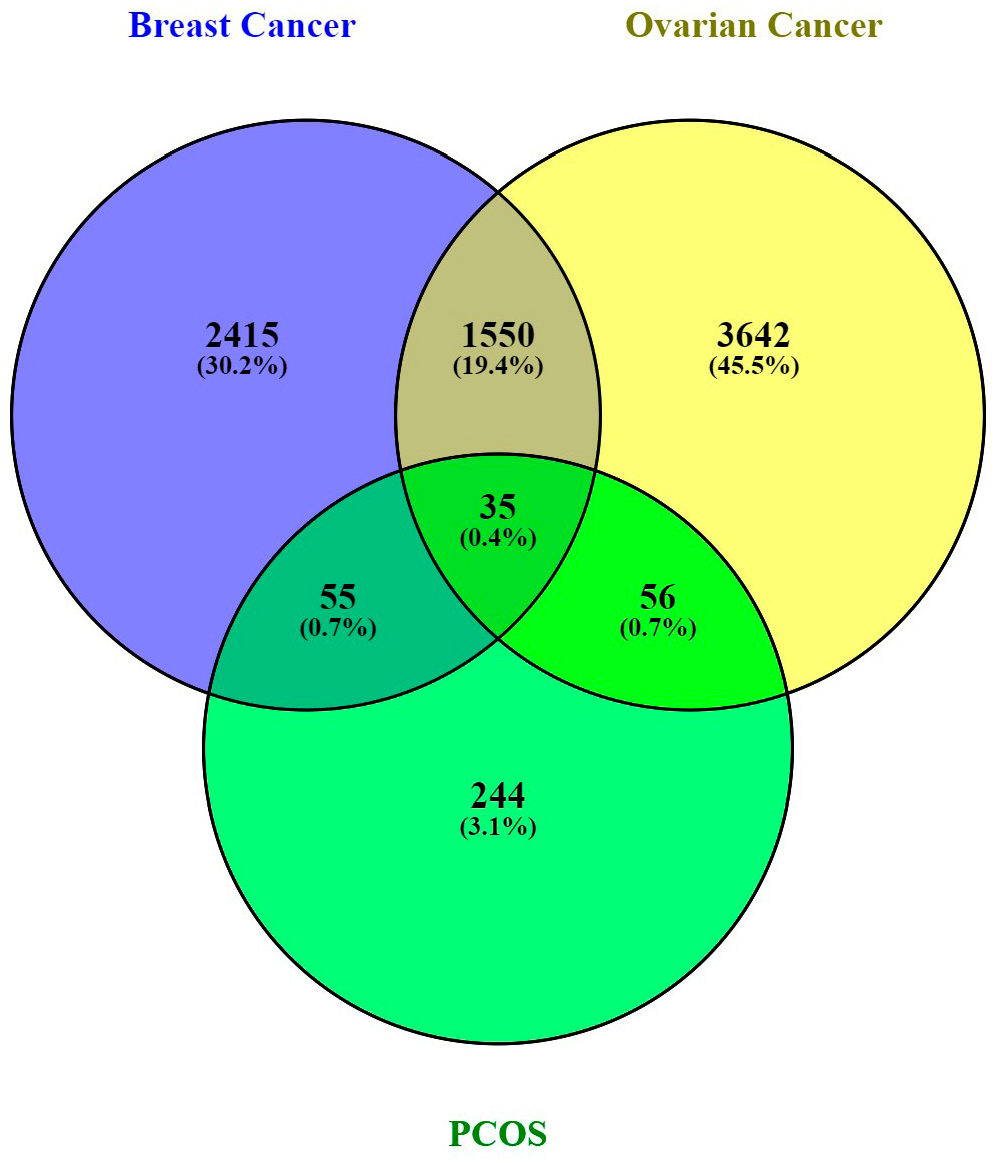
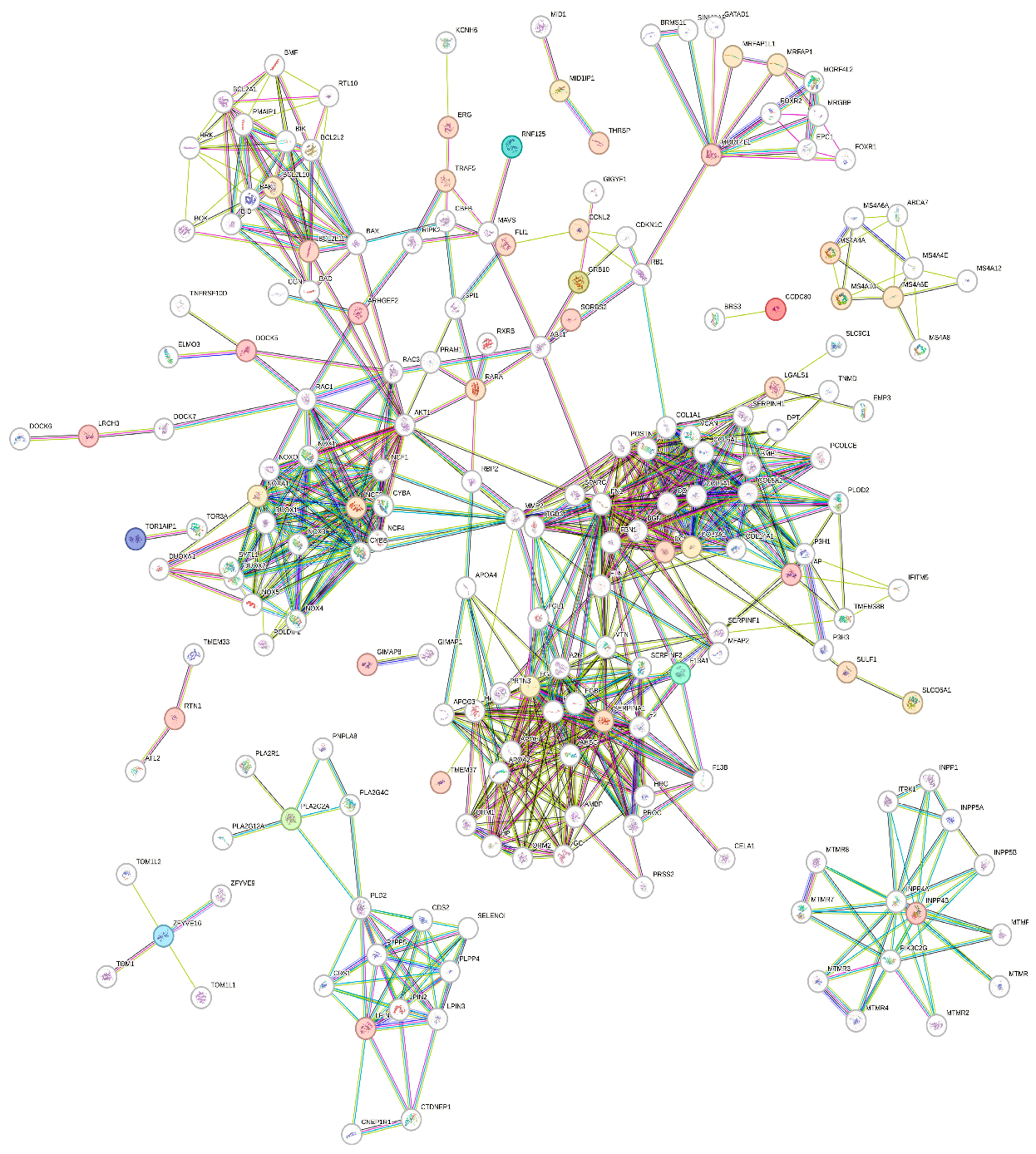
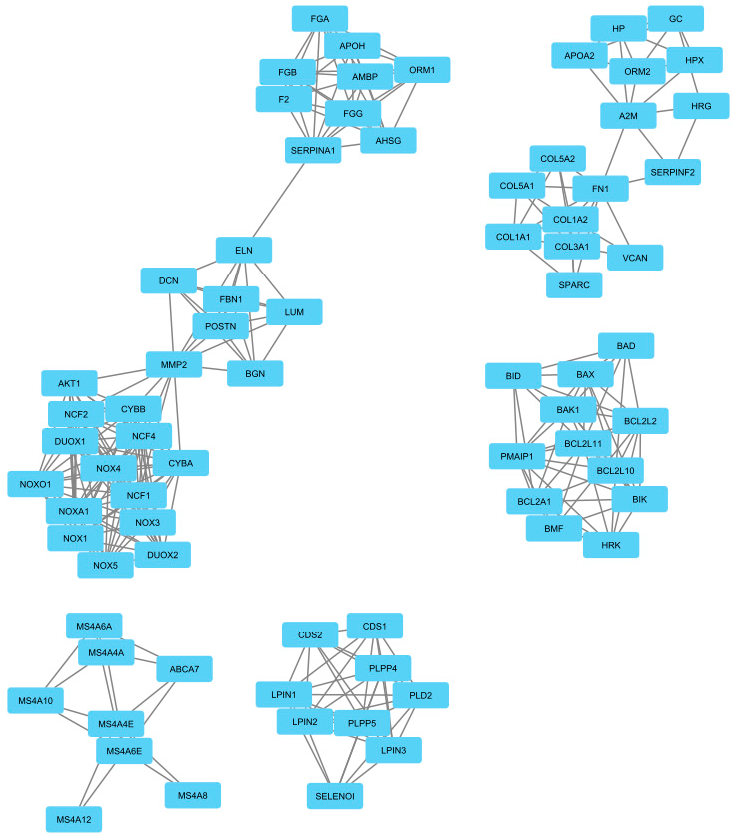
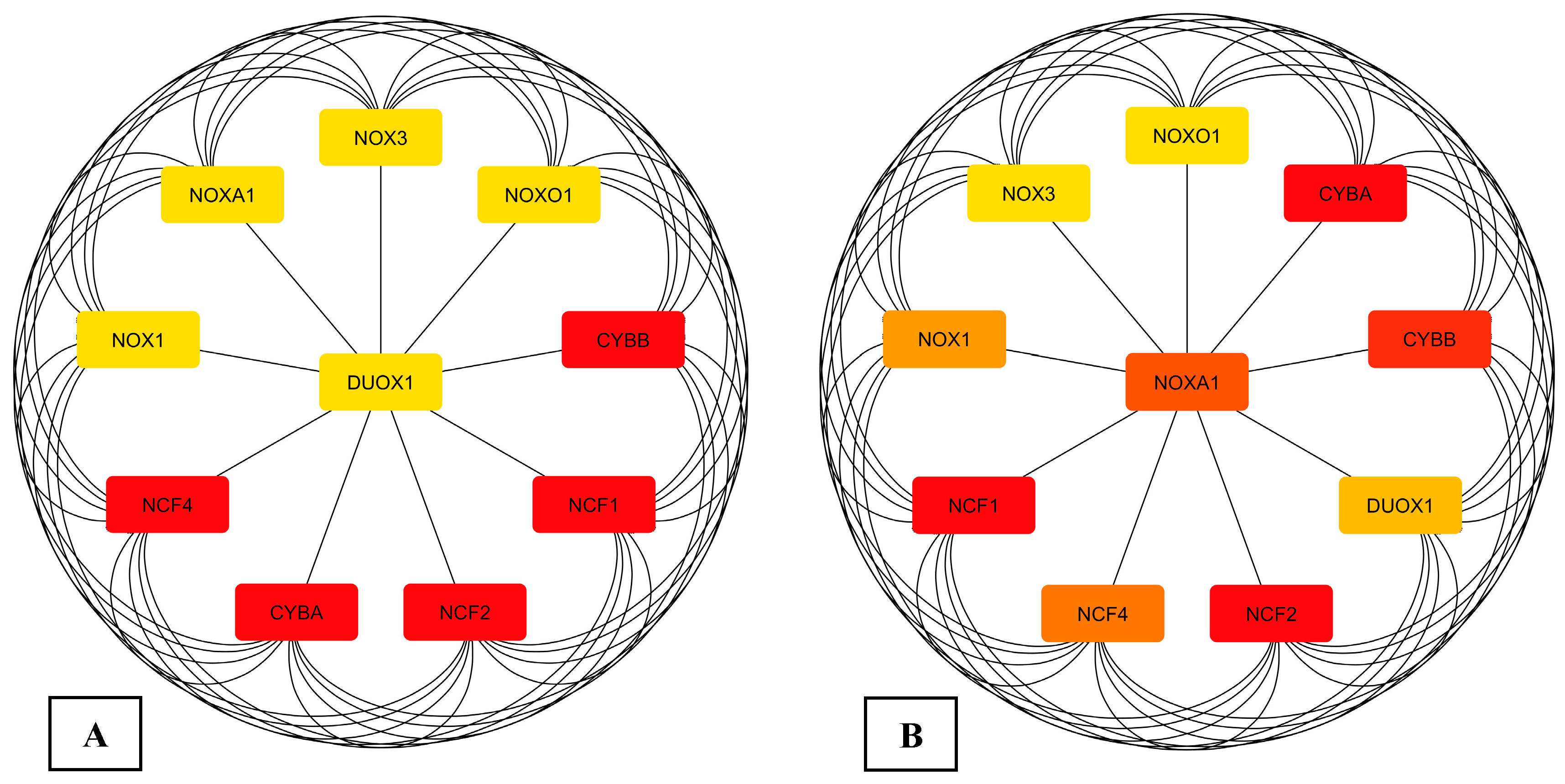
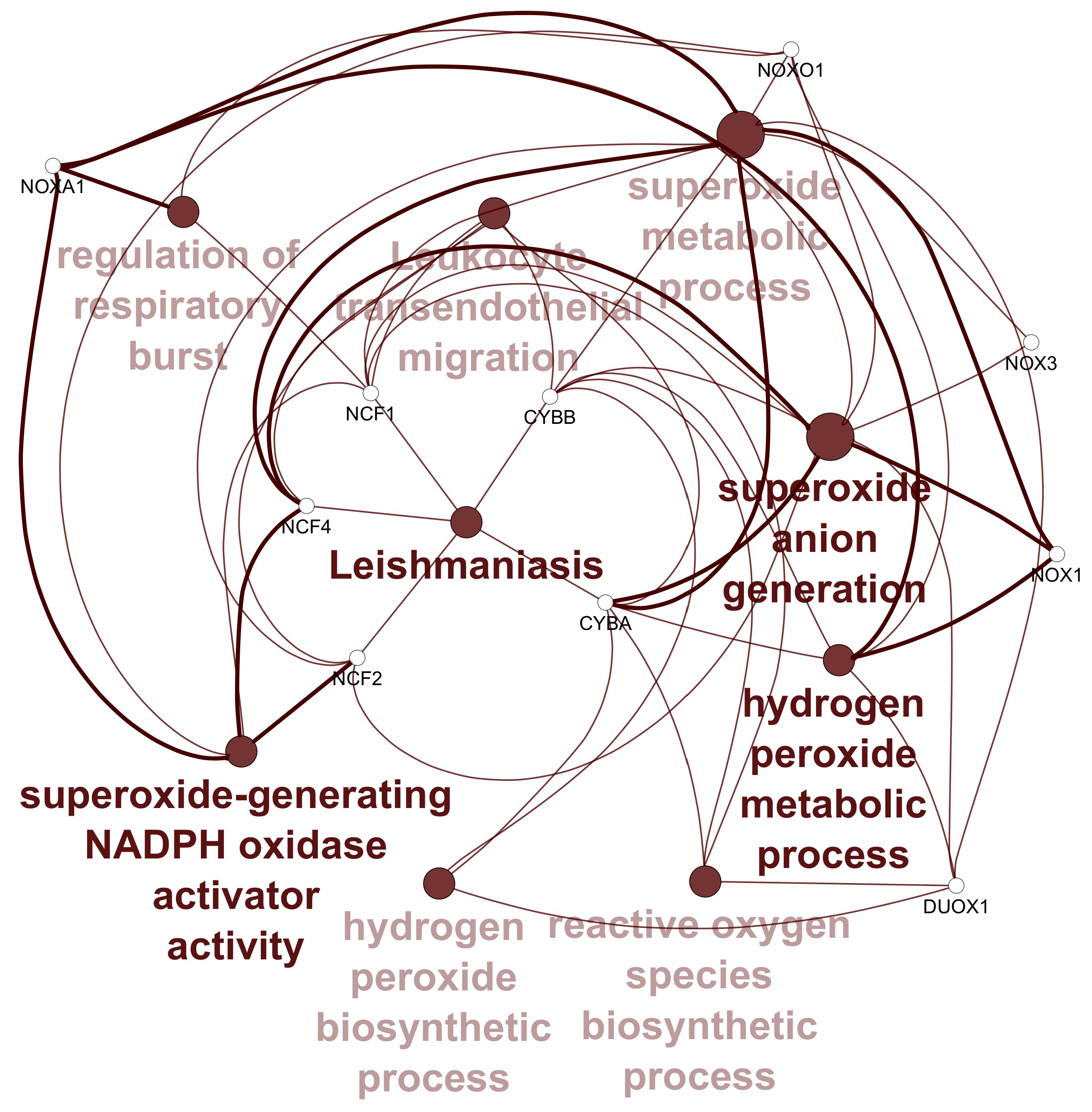
3.1. Data Retrieval and Identification of DEGs The datasets for PCOS, BC, and OC with series identifiers GSE84958, GSE42568, and GSE18520, respectively, were pre-processed to identify the DEGs by plotting volcano plots (Figure 2 and Table 1), followed by cross-comparison analysis that identified 35 common DEGs amongst PCOS, BC, and OC (Figure 3). Differentially Expressed Genes (DEGs) identified. 3.2. PPI Construction, Analysis, and Identification of Hub Genes An interactome representing protein-protein interactions of the 35 common DEGs shared amongst PCOS, BC, and OC was constructed in the STRING database at a high confidence value of 0.7 (indicates that the interaction between two proteins is more likely to be biologically relevant or experimentally validated). The resulting network comprised 195 nodes (representing genes) and 738 edges (representing interactions between nodes) (Figure 4). For identifying functional modules and predicting protein complexes in biological networks obtained from STRING, Cytoscape and its plugins MCODE and CytoCluster (ClusterONE algorithm) were used. On analysis of the interactome with MCODE, twelve clusters were obtained with five significant clusters based on the number of nodes and edges (Figure 5 and Table 2). Meanwhile, ClusterONE yielded twenty-four clusters, nine of which were significant, with a p-value < 0.05 (Table 3). The merged network of significant clusters of ClusterONE and MCODE was analyzed using the Cytoscape plugin CytoHubba, employing the Maximum Clique Centrality method (MCC). This analysis identified the top 10 hub genes, i.e., CYBA, CYBB, DUOX1, NCF1, NCF2, NCF4, NOX1, NOX3, NOXA1, and NOXO1 (Figure 6). The hub genes/nodes represent highly connected nodes having a higher likelihood to be involved in an essential interaction [48]. The hub genes were validated by analyzing the merged network of significant clusters of MCODE using the Cytoscape plugin NetworkAnalyzer, which computes and visualizes various directed and undirected network parameters [45]. The same hub genes were identified, and they were also found to be closely associated in the protein-protein interaction (PPI) network constructed and visualized using STRING (Table 4). Clusters selected from analysis of STRING network using Cytoscape plugin MCODE. Clusters selected from analysis of STRING network using Cytoscape plugin CytoCluster. Top 10 ranked hub genes based on degree obtained from analysis of MCODE clusters merged network using Cytoscape plug-in NetworkAnalyst and their interacting network in STRING. 3.3. Gene Set Enrichment Analysis All the gene products of the identified hub genes CYBA, CYBB, DUOX1, NCF1, NCF2, NCF4, NOX1, NOX3, NOXA1, and NOXO1 were enriched in reactive oxygen species metabolic processes, i.e., superoxide-generating NADPH oxidase activator activity (GO:0016176) and reactive oxygen species biosynthetic processes (GO:1903409), on analysis with Cytoscape plug-in Clue GO (Figure 7).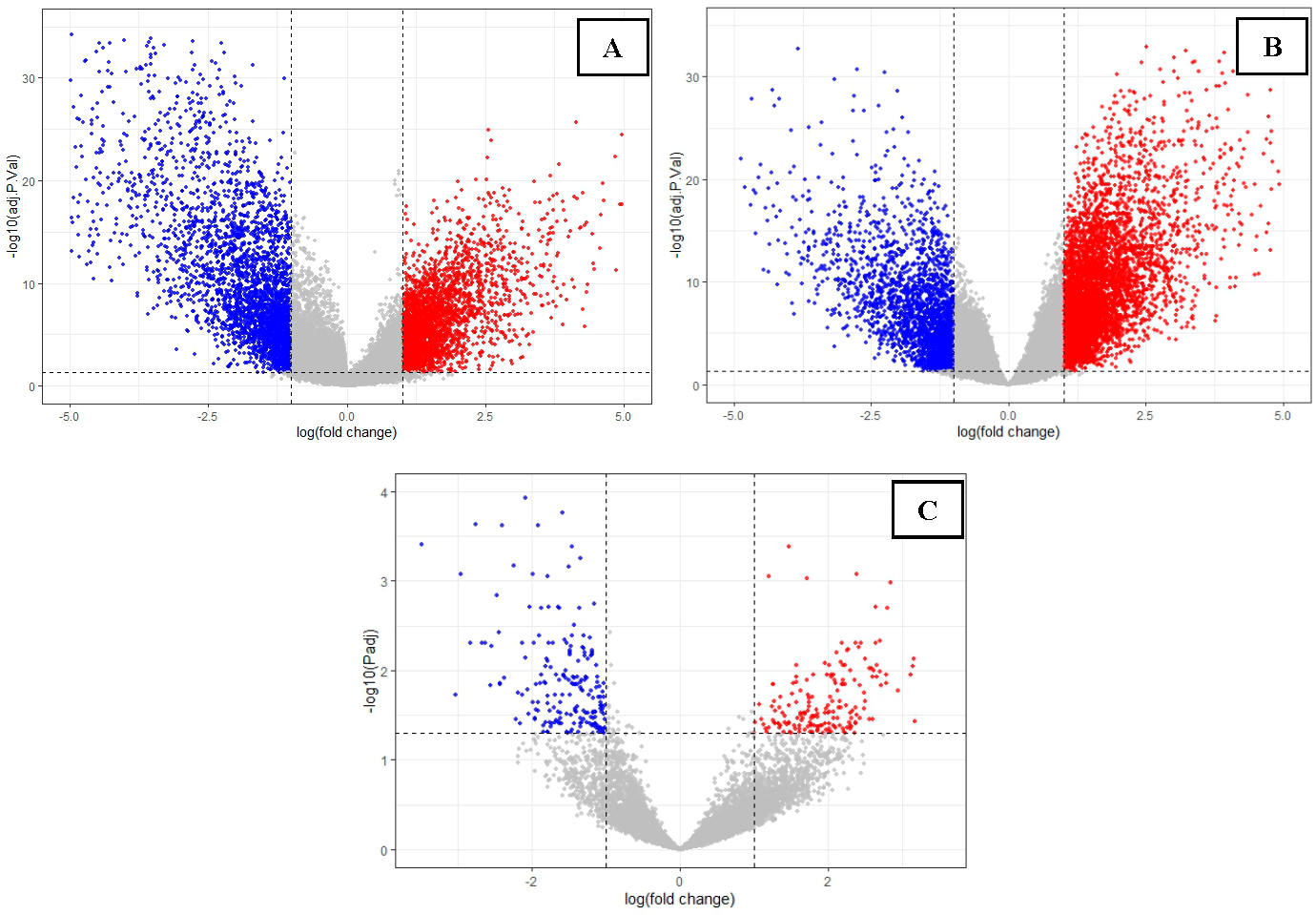
Disease
PCOS
Breast Cancer (BC)
Ovarian Cancer (OC)
No. of up-regulated genes
185
1909
3295
No. of down-regulated genes
205
2148
1988
Clusters
Nodes
Edges
Score
Clusters
Nodes
Edges
Score

30
151
10.4
12
50
9.1

9
34
8.5
16
41
5.5

8
16
4.571
Cluster
Details
Cluster
Details

Nodes: 25
Density: 0.550
Quality 0.846
P-value: 1.816E-9
Nodes: 28
Density: 0.447
Quality 0.790
P-value: 2.220E-9

Nodes: 18
Density: 0.725
Quality 0.847
P-value: 1.866E-7
Nodes: 14
Density: 0.648
Quality 0.937
P-value: 1.817E-6

Nodes: 11
Density: 0.709
Quality 0.951
P-value: 1.963E-5
Nodes: 11
Density: 0.509
Quality 0.800
P-value: 9.390E-5

Nodes: 8
Density: 0.571
Quality 1.000
P-value: 1.787E-4
Nodes: 9
Density: 0.500
Quality 0.857
P-value: 3.036E-4

Nodes: 10
Density: 0.533
Quality 0.667
P-value: 0.002
Gene

NCF1
NOX1
NCF2
NOX3
NCF4
NOXA1
CYBA
NOXO1
CYBB
DUOX1
The CYBA gene encodes the alpha subunit of cytochrome b-245 (also known as P22phox), while the CYBB/NOX2 gene provides instructions for producing the cytochrome b-245 protein. Beta chain (also known as p91phox), NCF1 (Neutrophil cytoplasmic factor 1/p47phox), NCF2 (Neutrophil cytoplasmic factor 2/pp67phox), and NCF4 (Neutrophil cytoplasmic factor 4 also known as p40phox) are components of NADPH oxidase required for its function [49]. p91phox (NOX2) is the catalytic subunit of the superoxide-generating respiratory burst NADPH oxidase, which is regulated by subunits p47phox and p67phox. A homolog of gp91phox, NOX1, is regulated by NOXO1, an organizer protein that cooperates with an activator protein, NOXA1, to regulate the catalytic subunit, both of which are homologs of p47phox and p67phox, respectively. DUOX1 (Dual oxidase 1) is also a member of the NADPH oxidase family that catalyzes the production of hydrogen peroxide and plays a crucial role in innate host defense and thyroid hormone biosynthesis [50]. In mammals, all NOX proteins produce ROS, with DUOX1–2 and NOX4 generating hydrogen peroxide, while others produce superoxide [51]. Studies have identified potential biomarkers and hub genes for breast and ovarian cancers through bioinformatics analyses. Common hub genes identified across studies include CCNE1, CCNB2, VEGFA, and PTEN, and the DEGs were found to be involved in critical pathways such as cell cycle, p53 signaling, and drug metabolism [32,52,53,54]. Several hub genes have been identified as potential therapeutic targets or prognostic markers, such as FN1, IL6, FOS, CCNG1, ADAMTS1, RPS9, RPL11, RPS14, and RPL10A. Key pathways involved in breast cancer development include cell adhesion, immune response, cell cycle, cell migration, proliferation, and p53 signaling [32,54,55,56]. FOS, CDKN1A, CD44, BCL2, and MMP3 have been implicated as being potentially important in ovarian cancer progression. Functional enrichment analysis revealed their involvement of these genes in processes such as collagen catabolism and stress-activated protein kinase cascades [57,58,59]. Notably, BUB1B and KIF20A were found to correlate with ovarian cancer prognosis and clinical characteristics [60]. Similarly, bioinformatics analyses have revealed key pathways and genes involved in PCOS development, including those related to interferon signaling, platelet activation, and lipid metabolism. Additionally, epigenetic alterations, such as DNA hypomethylation of genes involved in lipid and steroid synthesis, may contribute to hyperandrogenism in PCOS [61]. Key genes identified across studies include AR and STK11, which are involved in the AMPK and adipocytokine signaling pathways [62]. In another study, the identified DEGs were mainly involved in actin cytoskeleton organization, positive regulation of the NF-κB signaling pathway, and positive regulation of the canonical Wnt signaling pathway, along with the PI3K/Akt signaling pathway and glycosaminoglycan biosynthesis [33]. Recent studies have highlighted the role of increased oxidative stress, characterized by elevated ROS and reduced antioxidant activity in PCOS [32,63]. Polymorphism of cytokine genes, including IL1A, IL1B, IL1RN, 3and IL6, as well as genetic variations in NADPH oxidase components, have been linked to PCOS susceptibility [64,65,66]. Inhibition of NOX4 has been shown to reduce oxidative stress and cell apoptosis in PCOS rat models [32]. Additionally, inflammation-related genes have been reported to be differentially expressed in ovarian stroma and granulosa cells of PCOS women, suggesting alterations in the local ovarian immune system [67]. The imbalance in ROS distribution may contribute to reproductive dysfunction in PCOS, affecting follicular development, oocyte maturation, and ovulation [68]. This is the first study that reports a direct link between oxidative stress and PCOS, BC, and OC since all the identified common genes are components or regulators of the NADPH oxidase. NOX4, localized to mitochondria, plays a significant role in breast and ovarian cancer progression by generating ROS. It has been reported to contribute to oncogenic processes and also hyperandrogenism in PCOS [12,13,24]. Therefore, modulating NADPH oxidase activity could potentially serve as a clinically significant therapeutic approach to limit excessive inflammation in various diseases, as ROS and inflammation are closely interconnected, each capable of inducing the other. This could prevent the initiation and progression of breast and ovarian cancers in PCOS patients, as oxidative stress appears to be a crucial factor in PCOS pathophysiology, interacting with other etiological factors and environmental influences. Antioxidant supplementation and lifestyle modifications that restore oxidative balance and improve PCOS symptoms may further prevent the initiation, progression, and metastasis of cancers in females. The limitation of this study, which utilizes an interactome-based bioinformatics approach, is that although the role of the NOX gene family has been reported in multiple studies, the key pathways and hub genes identified in this study have not yet been validated through wet lab experiments or clinical studies. Second, due to the lack of a dataset of patients with combined conditions of PCOS and BC or OC, the validation of the hub gene was performed using in silico approaches and literature mining. Therefore, the results need to be confirmed by prospective clinical and wet lab studies.
In conclusion, this study explored the common hub genes and the possible biological mechanisms for the co-occurrence trend of PCOS, BC, and OC. The elevated oxidative stress in PCOS patients may increase their risk of developing cancer, especially breast and ovarian cancers, as free radicals promote carcinogenesis and metastasis. These findings highlight the complex interplay between the NOX gene family and ROS in PCOS, breast, and ovarian cancer and their potential as therapeutic targets, particularly for PCOS management.
ROS
Reactive Oxygen Species
PCOS
Polycystic Ovary Syndrome
DEGs
Differentially Expressed Genes
OS
Oxidative Stress
MDA
Malondialdehyde
GPX3
Glutathione Peroxidase 3
OC
Ovarian Cancer
BC
Breast Cancer
PPI
Protein-Protein Interactions
MCC
Maximum Clique Centrality Method
NCF1
Neutrophil Cytoplasmic Factor 1/p47phox
DUOX1
Dual Oxidase 1
The author is solely responsible for the conception, design, analysis, interpretation, drafting, and final approval of the article.
The datasets generated or analyzed during this study are available from the corresponding author upon reasonable request.
The study did not involve human subjects or animal models, and therefore, no ethical clearance was required.
Not applicable.
The author declares there is no conflicts of interest.
The study didn’t receive any funding.
The author would like acknowledge the assistance provided by Mr. Arnesh Saxena, post-graduate student at the Centre for Systems Biology and Bioinformatics, in analysis of the data sets. The author would also like to thank the parent institute, Panjab University Chandigarh, India, for providing the infrastructure for carrying out the research.
[1] Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [CrossRef] [PubMed]
[2] Madkour, L.H. . Reactive Oxygen Species (ROS), Nanoparticles, and Endoplasmic Reticulum (ER) Stress-Induced Cell Death Mechanisms ; Academic Press: Cambridge, MA, USA, 2020; . .
[3] de Araújo, R.F.; Martins, D.B.; Borba, M.A. . Oxidative Stress and Disease. InA Master Regulator of Oxidative Stress-the Transcription Factor nrf2 ; IntechOpen: London, UK, 2016; . .
[4] Noda, N.; Wakasugi, H. Cancer and Oxidative Stress. Jpn. Med. Assoc. J. 2001, 44, 535–539.
[5] Visconti, R.; Grieco, D. New Insights on Oxidative Stress in Cancer. Curr. Opin. Drug Discov. Dev. 2009, 12, 240–245.
[6] Nourazarian, A.R.; Kangari, P.; Salmaninejad, A. Roles of Oxidative Stress in the Development and Progression of Breast Cancer. Asian Pac. J. Cancer Prev. 2014, 15, 4745–4751. [CrossRef]
[7] Zhang, S.; Xiao, X.; Yi, Y.; Wang, X.; Zhu, L.; Shen, Y.; Lin, D.; Wu, C. Tumor Initiation and Early Tumorigenesis: Molecular Mechanisms and Interventional Targets. Signal Transduct. Target. Ther. 2024, 9, 149. [CrossRef]
[8] Neganova, M.; Liu, J.; Aleksandrova, Y.; Klochkov, S.; Fan, R. Therapeutic Influence on Important Targets Associated with Chronic Inflammation and Oxidative Stress in Cancer Treatment. Cancers 2021, 13. [CrossRef] [PubMed]
[9] Hecht, F.; Pessoa, C.F.; Gentile, L.B.; Rosenthal, D.; Carvalho, D.P.; Fortunato, R.S. The Role of Oxidative Stress on Breast Cancer Development and Therapy. Tumor Biol. 2016, 37, 4281–4291. [CrossRef]
[10] Saed, G.M.; Diamond, M.P.; Fletcher, N.M. Updates of the Role of Oxidative Stress in the Pathogenesis of Ovarian Cancer. Gynecol. Oncol. 2017, 145, 595–602. [CrossRef]
[11] Souza, A.d.V.e.; de Faria, C.C.; Pereira, L.M.; Ferreira, A.C.F.; Torres, P.H.M.; Fortunato, R.S. Gene Expression and Prognostic Value of NADPH Oxidase Enzymes in Breast Cancer. Int. J. Mol. Sci. 2024, 25. [CrossRef]
[12] Desouki, M.M.; Kulawiec, M.; Bansal, S.; Das, G.C.; Singh, K.K. Cross Talk Between Mitochondria and Superoxide Generating NADPH Oxidase in Breast and Ovarian Tumors. Cancer Biol. Ther. 2005, 4, 1367–1373. [CrossRef]
[13] Graham, K.A.; Kulawiec, M.; Owens, K.M.; Li, X.; Desouki, M.M.; Chandra, D.; Singh, K.K. NADPH Oxidase 4 Is an Oncoprotein Localized to Mitochondria. Cancer Biol. Ther. 2010, 10, 223–231. [CrossRef] [PubMed]
[14] Szanto, I. NADPH Oxidase 4 (NOX4) in Cancer: Linking Redox Signals to Oncogenic Metabolic Adaptation. Int. J. Mol. Sci. 2022, 23. [CrossRef] [PubMed]
[15] Cohen, S.; Mehrabi, S.; Yao, X.; Millingen, S.; Aikhionbare, F.O. Reactive Oxygen Species and Serous Epithelial Ovarian Adenocarcinoma. Cancer Res. J. 2016, 4, 106–114. [CrossRef]
[16] Singh, R.; Manna, P.P. Reactive Oxygen Species in Cancer Progression and Its Role in Therapeutics. Explor. Med. 2022, 3, 43–57. [CrossRef]
[17] Chen, Q.; Wang, Q.; Zhu, J.; Xiao, Q.; Zhang, L. Reactive Oxygen Species: Key Regulators in Vascular Health and Diseases. Br. J. Pharmacol. 2017, 175, 1279–1292. [CrossRef] [PubMed]
[18] Banerjee, S.; Lo, W.-C.; Majumder, P.; Roy, D.; Ghorai, M.; Shaikh, N.K.; Kant, N.; Shekhawat, M.S.; Gadekar, V.S.; Ghosh, S.; et al. Multiple Roles for Basement Membrane Proteins in Cancer Progression and EMT. Eur. J. Cell Biol. 2022, 101. [CrossRef]
[19] Das, A.; Roychoudhury, S. Reactive Oxygen Species in the Reproductive System: Sources and Physiological Roles. Oxidative Stress and Toxicity in Reproductive Biology and Medicine: A Comprehensive Update on Male Infertility-Volume One ; Springer International Publishing: Cham, Switzerland, 2022; 9–40. .
[20] Rizzo, A.; Roscino, M.; Binetti, F.; Sciorsci, R. Roles of Reactive Oxygen Species in Female Reproduction. Reprod. Domest. Anim. 2011, 47, 344–352. [CrossRef]
[21] Abed, S.K.; Jasim, H.H.; Shahooth, M.A.; Saud, M.A. The Association Between Free Radicals and the Ovarian Functions: A Review. Al-Anbar J. Vet.-Sci. 2024, 17, 11–17. [CrossRef]
[22] Rudnicka, E.; Duszewska, A.M.; Kucharski, M.; Tyczyński, P.; Smolarczyk, R. Oxidative Stress and Reproductive Function: Oxidative Stress in Polycystic Ovary Syndrome. Reproduction 2022, 164, F145–F154. [CrossRef]
[23] Azziz, R. Polycystic Ovary Syndrome. Obstet. Gynecol. 2018, 132, 321–336. [CrossRef]
[24] González, F. Inflammation in Polycystic Ovary Syndrome: Underpinning of Insulin Resistance and Ovarian Dysfunction. Steroids 2011, 77, 300–305. [CrossRef] [PubMed]
[25] Sengupta, P.; Dutta, S.; Hassan, M.F. Polycystic Ovary Syndrome (PCOS) and Oxidative Stress. J. Integr. Sci. Technol. 2024, 12, 752–752. [CrossRef]
[26] Shen, S.-H.; Liou, T.-H.; Hsu, M.-I.; Chang, Y.-C.I.; Cheng, C.-Y.; Hsu, C.-S.; Tzeng, C.-R. Obesity and Inflammatory Biomarkers in Women with Polycystic Ovary Syndrome. Eur. J. Obstet. Gynecol. Reprod. Biol. 2015, 192, 66–71. [CrossRef]
[27] Modi, K.B. Poly Cystic Ovarian Syndrome (PCOS) and Gynecological Cancers. Med. J. 2024, II, 80. [CrossRef]
[28] Simic, P.; Coric, V.; Pljesa, I.; Savic-Radojevic, A.; Zecevic, N.; Kocic, J.; Simic, T.; Pazin, V.; Pljesa-Ercegovac, M. The Role of Glutathione Transferase Omega-Class Variant Alleles in Individual Susceptibility to Ovarian Cancer. Int. J. Mol. Sci. 2024, 25. [CrossRef] [PubMed]
[29] Barry, J.A.; Azizia, M.M.; Hardiman, P.J. Risk of Endometrial, Ovarian and Breast Cancer in Women with Polycystic Ovary Syndrome: A Systematic Review and Meta-Analysis. Hum. Reprod. Updat. 2014, 20, 748–758. [CrossRef]
[30] Zuo, T.; Zhu, M.; Xu, W. Roles of Oxidative Stress in Polycystic Ovary Syndrome and Cancers. Oxidative Med. Cell. Longev. 2015, 2016, 8589318. [CrossRef]
[31] Harris, H.R.; Terry, K.L. Polycystic Ovary Syndrome and Risk of Endometrial, Ovarian, And Breast Cancer: A Systematic Review. Fertil. Res. Pr. 2016, 2, 14. [CrossRef]
[32] Li, Z.; Wang, Y.-H.; Wang, L.-L.; Hu, D.-T.; Teng, Y.; Zhang, T.-Y.; Yan, Z.-Y.; Wang, F.; Zou, Y.-F. Polycystic Ovary Syndrome and the Risk of Endometrial, Ovarian and Breast Cancer: An Updated Meta-Analysis. Scott. Med. J. 2022, 67, 109–120. [CrossRef]
[33] Zhou, J.; Jiang, Z.; Fu, L.; Qu, F.; Dai, M.; Xie, N.; Zhang, S.; Wang, F. Contribution of Labor Related Gene Subtype Classification on Heterogeneity of Polycystic Ovary Syndrome. PLoS ONE 2023, 18. [CrossRef]
[34] Tuly, K.F.; Hossen, M.B.; Islam, M.A.; Kibria, M.K.; Alam, M.S.; Harun-Or-Roshid, M.; Begum, A.A.; Hasan, S.; Mahumud, R.A.; Mollah, M.N. Robust Identification of Differential Gene Expression Patterns from Multiple Transcriptomics Datasets for Early Diagnosis, Prognosis, and Therapies for Breast Cancer. Medicina 2023, 59. [CrossRef]
[35] Zuo, L.; Li, X.; Tan, Y.; Zhu, H.; Xiao, M. Prospective Pathway Signaling and Prognostic Values of MicroRNA-9 in Ovarian Cancer Based on Gene Expression Omnibus (GEO): A Bioinformatics Analysis. J. Ovarian Res. 2021, 14, 29. [CrossRef] [PubMed]
[36] Davis, S.; Meltzer, P.S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007, 23, 1846–1847. [CrossRef]
[37] Kolberg, L.; Raudvere, U.; Kuzmin, I.; Vilo, J.; Peterson, H. gprofiler2--an R Package for gene List Functional Enrichment Analysis and Namespace Conversion Toolset g: Profiler. F1000Research 2020, 9, ELIXIR-709. [CrossRef]
[38] Tyner, S.; Briatte, F.; Hofmann, H. Network Visualization with ggplot2. R J. 2017, 9, 27–59. [CrossRef]
[39] Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [CrossRef] [PubMed]
[40] Oliveros, J.C. VENNY. An Interactive Tool for Comparing Lists with Venn Diagrams. 2007 Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 14 September 2024).
[41] Mering, C.V.; Huynen, M.; Jaeggi, D.; Schmidt, S.; Bork, P.; Snel, B. STRING: A Database of Predicted Functional Associations Between Proteins. Nucleic Acids Res. 2003, 31, 258–261. [CrossRef]
[42] Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [CrossRef] [PubMed]
[43] Li, M.; Li, D.; Tang, Y.; Wu, F.; Wang, J. CytoCluster: A Cytoscape Plugin for Cluster Analysis and Visualization of Biological Networks. Int. J. Mol. Sci. 2017, 18. [CrossRef]
[44] Bader, G.D.; Hogue, C.W.V. An Automated Method for Finding Molecular Complexes in large Protein Interaction Networks. BMC Bioinform. 2003, 4, 2–27. [PubMed] [CrossRef]
[45] Assenov, Y.; Ramírez, F.; Schelhorn, S.-E.; Lengauer, T.; Albrecht, M. Computing Topological Parameters of Biological Networks. Bioinformatics 2008, 24, 282–284. [CrossRef] [PubMed]
[46] Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying Hub Objects and Sub-Networks from Complex Interactome. BMC Syst. Biol. 2014, 8, 1–7. [CrossRef] [PubMed]
[47] Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape Plug-in to Decipher Functionally Grouped Gene Ontology and Pathway Annotation Networks. Bioinformatics 2009, 25, 1091–1093. [CrossRef] [PubMed]
[48] Sahrawat, T.R.; Dwivedi, J. Investigating the Relationship between Diabetes and Alzheimer’s Disease:: A Network Systems Biology Approach. J. Sci. Sustain. Dev. 2020, 7. [CrossRef]
[49] Cheng, G.; Ritsick, D.; Lambeth, J.D. Nox3 Regulation by NOXO1, p47phox, and p67phox. J. Biol. Chem. 2004, 279, 34250–34255. [CrossRef]
[50] De Deken, X.; Corvilain, B.; Dumont, J.E.; Miot, F. Roles of DUOX-Mediated Hydrogen Peroxide in Metabolism, Host Defense, and Signaling. Antioxid. Redox Signal. 2014, 20, 2776–2793. [CrossRef]
[51] Brandes, R.P.; Weissmann, N.; Schröder, K. Nox Family NADPH Oxidases: Molecular Mechanisms of Activation. Free Radic. Biol. Med. 2014, 76, 208–226. [CrossRef]
[52] Zou, J.; Li, Y.; Liao, N.; Liu, J.; Zhang, Q.; Luo, M.; Xiao, J.; Chen, Y.; Wang, M.; Chen, K.; et al. Identification of Key Genes Associated with Polycystic Ovary Syndrome (PCOS) and Ovarian Cancer Using an Integrated Bioinformatics Analysis. J. Ovarian Res. 2022, 15, 30. [CrossRef]
[53] Atasever, S. Identification of Potential Hub Genes as Biomarkers for Breast, Ovarian, and Endometrial Cancers. Front. Life Sci. Relat. Technol. 2024, 5, 74–82. [CrossRef]
[54] Fang, E.; Zhang, X. Identification of Breast Cancer Hub Genes and Analysis of Prognostic Values Using Integrated Bioinformatics Analysis. Cancer Biomark. 2018, 21, 373–381. [CrossRef]
[55] Jin, H.; Huang, X.; Shao, K.; Li, G.; Wang, J.; Yang, H.; Hou, Y. Integrated Bioinformatics Analysis to Identify 15 Hub Genes in Breast Cancer. Oncol. Lett. 2019, 18, 1023–1034. [CrossRef]
[56] Hao, M.; Liu, W.; Ding, C.; Peng, X.; Zhang, Y.; Chen, H.; Dong, L.; Liu, X.; Zhao, Y.; Chen, X.; et al. Identification of Hub Genes and Small Molecule Therapeutic Drugs Related to Breast Cancer with Comprehensive Bioinformatics Analysis. PeerJ 2020, 8, e9946. [CrossRef] [PubMed]
[57] Zhao, Y.; Pi, J.; Liu, L.; Yan, W.; Ma, S.; Hong, L. Identification of the Hub Genes Associated with the Prognosis of Ovarian Cancer Patients via Integrated Bioinformatics Analysis and Experimental Validation. Cancer Manag. Res. 2021, 13, 707–721. [CrossRef] [PubMed]
[58] Zhou, X.; Song, Z.; Chen, J.; Wang, D.; Sun, J. Identification of Potential Hub Genes and Therapeutic Drugs in Ovarian Cancer via Bioinformatics Analysis. All Life 2023, 16, 2270192. [CrossRef]
[59] Yin, J.-G.; Liu, X.-Y.; Wang, B.; Wang, D.-Y.; Wei, M.; Fang, H.; Xiang, M. Gene Expression Profiling Analysis of Ovarian Cancer. Oncol. Lett. 2016, 12, 405–412. [CrossRef] [PubMed]
[60] Zhang, J.; Yuan, B.; Zhang, H.; Li, H. Human Epithelial Ovarian Cancer Cells Expressing CD105, CD44 and CD106 Surface Markers Exhibit Increased Invasive Capacity and Drug Resistance. Oncol. Lett. 2019, 17, 5351–5360. [CrossRef]
[61] Pan, J.-X.; Tan, Y.-J.; Wang, F.-F.; Hou, N.-N.; Xiang, Y.-Q.; Zhang, J.-Y.; Liu, Y.; Qu, F.; Meng, Q.; Xu, J.; et al. Aberrant Expression and DNA Methylation of Lipid Metabolism Genes in PCOS: A New Insight into Its Pathogenesis. Clin. Epigenet. 2018, 10, 6. [CrossRef]
[62] Gollapalli, P.; Kumari, N.S.; Shetty, P.; Gnanasekaran, T.S. Molecular Basis of AR and STK11 Genes Associated Pathogenesis via AMPK Pathway and Adipocytokine Signalling Pathway in the Development of Metabolic Disorders in PCOS Women. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 23. [CrossRef]
[63] Krishna, M.B.; Joseph, A.; Thomas, P.L.; Dsilva, B.; Pillai, S.M.; Laloraya, M. Impaired Arginine Metabolism Coupled to a Defective Redox Conduit Contributes to Low Plasma Nitric Oxide in Polycystic Ovary Syndrome. Cell. Physiol. Biochem. 2017, 43, 1880–1892. [CrossRef]
[64] Ma, W.; Li, S.; Liu, H.; Bai, H.; Liu, Q.; Hu, K.; Guan, L.; Fan, P. Myeloperoxidase and CYBA Genetic Variants in Polycystic Ovary Syndrome. Eur. J. Clin. Investig. 2021, 51, e13438. [CrossRef]
[65] Stasia, M.J. CYBA Encoding p22phox, the Cytochrome b558 Alpha Polypeptide: Gene Structure, Expression, Role and Physiopathology. Gene 2016, 586, 27–35. [CrossRef]
[66] de Alencar, J.B.; Alves, H.V.; Elpidio, L.N.S.; Visentainer, J.E.L.; Sell, A.M. Polymorphisms of Cytokine Genes and Polycystic Ovary Syndrome: A Review. Metab. Syndr. Relat. Disord. 2016, 14, 468–474. [CrossRef] [PubMed]
[67] Schmidt, J.; Weijdegard, B.; Mikkelsen, A.L.; Lindenberg, S.; Nilsson, L.; Brannstrom, M. Differential Expression of Inflammation-Related Genes in the Ovarian Stroma and Granulosa Cells of PCOS Women. Mol. Hum. Reprod. 2013, 20, 49–58. [CrossRef] [PubMed]
[68] Awonuga, A.O.; Camp, O.G.; Abu-Soud, H.M. A Review of Nitric Oxide and Oxidative Stress in Typical Ovulatory Women and in the Pathogenesis of Ovulatory Dysfunction in PCOS. Reprod. Biol. Endocrinol. 2023, 21. [CrossRef] [PubMed]
We use cookies to improve your experience on our site. By continuing to use our site, you accept our use of cookies. Learn more